Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis
- PMID: 28426876
- PMCID: PMC5862314
- DOI: 10.1093/carcin/bgx005
Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis
Abstract
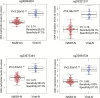
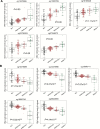
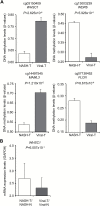
The aim of this study was to clarify the significance of DNA methylation alterations during non-alcoholic steatohepatitis (NASH)-related hepatocarcinogenesis. Single-CpG-resolution genome-wide DNA methylation analysis was performed on 264 liver tissue samples using the Illumina Infinium HumanMethylation450 BeadChip. After Bonferroni correction, 3331 probes showed significant DNA methylation alterations in 113 samples of non-cancerous liver tissue showing NASH (NASH-N) as compared with 55 samples of normal liver tissue (NLT). Principal component analysis using the 3331 probes revealed distinct DNA methylation profiles of NASH-N samples that were different from those of NLT samples and 37 samples of non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with hepatitis B virus (HBV) or hepatitis C virus (HCV) infection (viral-N). Receiver operating characteristic curve analysis identified 194 probes that were able to discriminate NASH-N samples from viral-N samples with area under the curve values of more than 0.95. Jonckheere-Terptsra trend test revealed that DNA methylation alterations in NASH-N samples from patients without hepatocellular carcinoma (HCC) were inherited by or strengthened in NASH-N samples from patients with HCC, and then inherited by or further strengthened in 22 samples of NASH-related HCC (NASH-T) themselves. NASH- and NASH-related HCC-specific DNA methylation alterations, which were not evident in viral-N samples and 37 samples of HCC associated with HBV or HCV infection, were observed in tumor-related genes, such as WHSC1, and were frequently associated with mRNA expression abnormalities. These data suggested that NASH-specific DNA methylation alterations may participate in NASH-related multistage hepatocarcinogenesis.
© The Author 2017. Published by Oxford University Press.
Figures


Similar articles
-
Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas.J Cancer Res Clin Oncol. 2020 Oct;146(10):2461-2477. doi: 10.1007/s00432-020-03298-4. Epub 2020 Jul 19. J Cancer Res Clin Oncol. 2020. PMID: 32685988 Free PMC article.
-
Quantification of DNA methylation for carcinogenic risk estimation in patients with non-alcoholic steatohepatitis.Clin Epigenetics. 2022 Dec 5;14(1):168. doi: 10.1186/s13148-022-01379-4. Clin Epigenetics. 2022. PMID: 36471401 Free PMC article.
-
Comparison of genome-scale DNA methylation profiles in hepatocellular carcinoma by viral status.Epigenetics. 2016 Jun 2;11(6):464-74. doi: 10.1080/15592294.2016.1151586. Epub 2016 Jun 1. Epigenetics. 2016. PMID: 27248055 Free PMC article.
-
Molecular pathological approach to cancer epigenomics and its clinical application.Pathol Int. 2024 Apr;74(4):167-186. doi: 10.1111/pin.13418. Epub 2024 Mar 14. Pathol Int. 2024. PMID: 38482965 Free PMC article. Review.
-
Hepatocellular carcinoma and non-alcoholic steatohepatitis: The state of play.World J Gastroenterol. 2016 Feb 28;22(8):2494-502. doi: 10.3748/wjg.v22.i8.2494. World J Gastroenterol. 2016. PMID: 26937137 Free PMC article. Review.
Cited by
-
Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas.J Cancer Res Clin Oncol. 2020 Oct;146(10):2461-2477. doi: 10.1007/s00432-020-03298-4. Epub 2020 Jul 19. J Cancer Res Clin Oncol. 2020. PMID: 32685988 Free PMC article.
-
F-box protein 43 promoter methylation as a novel biomarker for hepatitis B virus-associated hepatocellular carcinoma.Front Microbiol. 2023 Nov 2;14:1267844. doi: 10.3389/fmicb.2023.1267844. eCollection 2023. Front Microbiol. 2023. PMID: 38029156 Free PMC article.
-
Quantification of DNA methylation for carcinogenic risk estimation in patients with non-alcoholic steatohepatitis.Clin Epigenetics. 2022 Dec 5;14(1):168. doi: 10.1186/s13148-022-01379-4. Clin Epigenetics. 2022. PMID: 36471401 Free PMC article.
-
Accumulation of Genetic and Epigenetic Alterations in the Background Liver and Emergence of Hepatocellular Carcinoma in Patients with Non-Alcoholic Fatty Liver Disease.Cells. 2021 Nov 21;10(11):3257. doi: 10.3390/cells10113257. Cells. 2021. PMID: 34831479 Free PMC article.
-
Methylation pattern of urinary DNA as a marker of kidney function decline in diabetes.BMJ Open Diabetes Res Care. 2020 Sep;8(1):e001501. doi: 10.1136/bmjdrc-2020-001501. BMJ Open Diabetes Res Care. 2020. PMID: 32883689 Free PMC article.
References
-
- Arai E., et al. (2010) DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics, 2, 467–481. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

