Identification of DBCCR1 as a suppressor in the development of lung cancer that is associated with increased DNA methyltransferase 1
- PMID: 28427182
- PMCID: PMC5464830
- DOI: 10.18632/oncotarget.15826
Identification of DBCCR1 as a suppressor in the development of lung cancer that is associated with increased DNA methyltransferase 1
Abstract
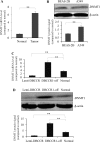
Accumulating evidence has pointed to a role of the CpG island hypermethylation in the regulation of cancer-related genes in tumor progression. However, the biological impacts in cancer pathogenesis associated with down-regulation of such gene targets remains elusive. Here we focused on a potential target of hypermethylation, DBCCR1 (deleted in bladder cancer chromosome region 1), a gene encoding a candidate tumor suppressor. We found that the expression of DBCCR1 is significantly lower in the lung cancer tissues compared with adjacent non-tumor tissues of patients. Importantly, the decreased DBCCR1 was found correlated with more advanced stages of cancer, and with a significantly shorter survival of patients. Genetic silencing DBCCR1 in human lung cancer cell line A549 resulted in an enhanced proliferation, migration, and invasion capacity. Conversely, restoring DBCCR1 expression blocked the growth and inhibited the ability of cancer cell in migration and invasion. Interestingly, DBCCR1 attenuates the expression of DNMT1 (DNA methyltransferase 1), suggesting a reciprocal regulation between genetic silencing of cancer suppressor genes and activating DNA methylation. Our data thus implicates DBCCR1 downregulation as a potential module in the pathogenesis of lung cancer through DNA methylation.
Keywords: DBCCR1; DNA methyltransferase; epigenetics; lung cancer; tumor suppressor.
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Figures




Similar articles
-
Methylation of RILP in lung cancer promotes tumor cell proliferation and invasion.Mol Cell Biochem. 2021 Feb;476(2):853-861. doi: 10.1007/s11010-020-03950-0. Epub 2020 Oct 30. Mol Cell Biochem. 2021. PMID: 33128214
-
Structure and methylation-based silencing of a gene (DBCCR1) within a candidate bladder cancer tumor suppressor region at 9q32-q33.Genomics. 1998 Mar 15;48(3):277-88. doi: 10.1006/geno.1997.5165. Genomics. 1998. PMID: 9545632
-
Loss of heterozygosity at 9q33 and hypermethylation of the DBCCR1 gene in oral squamous cell carcinoma.Br J Cancer. 2004 Aug 16;91(4):760-4. doi: 10.1038/sj.bjc.6601980. Br J Cancer. 2004. PMID: 15226771 Free PMC article.
-
Interplay between DNA Methyltransferase 1 and microRNAs During Tumorigenesis.Curr Drug Targets. 2021;22(10):1129-1148. doi: 10.2174/1389450122666210120141546. Curr Drug Targets. 2021. PMID: 33494674 Review.
-
Tumour suppression through modulation of neprilysin signaling: A comprehensive review.Eur J Pharmacol. 2021 Jan 15;891:173727. doi: 10.1016/j.ejphar.2020.173727. Epub 2020 Nov 5. Eur J Pharmacol. 2021. PMID: 33160935 Review.
Cited by
-
Epigenetic inactivation of HOXD10 is associated with human colon cancer via inhibiting the RHOC/AKT/MAPK signaling pathway.Cell Commun Signal. 2019 Jan 25;17(1):9. doi: 10.1186/s12964-018-0316-0. Cell Commun Signal. 2019. PMID: 30683109 Free PMC article.
-
Gene Targets Network Analysis for the Revealing and Guidance of Molecular Driving Mechanism of Lung Cancer.Front Genet. 2021 Sep 20;12:727201. doi: 10.3389/fgene.2021.727201. eCollection 2021. Front Genet. 2021. PMID: 34616430 Free PMC article.
-
Regulation of HAND2 Expression by LncRNA HAND2-AS1 in Ovarian Endometriosis Involving DNA Methylation.J Endocr Soc. 2023 Apr 18;7(6):bvad049. doi: 10.1210/jendso/bvad049. eCollection 2023 May 5. J Endocr Soc. 2023. PMID: 37153110 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
-
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. - PubMed
-
- Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur J Cancer. 2000;36:2294–2300. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

