Karyotypic evolution of the Medicago complex: sativa-caerulea-falcata inferred from comparative cytogenetic analysis
- PMID: 28427346
- PMCID: PMC5399346
- DOI: 10.1186/s12862-017-0951-x
Karyotypic evolution of the Medicago complex: sativa-caerulea-falcata inferred from comparative cytogenetic analysis
Abstract
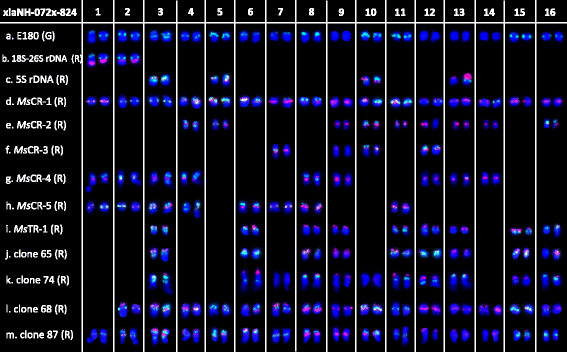
Background: Polyploidy plays an important role in the adaptation and speciation of plants. The alteration of karyotype is a significant event during polyploidy formation. The Medicago sativa complex includes both diploid (2n = 2× = 16) and tetraploid (2n = 2× = 32) subspecies. The tetraploid M. ssp. sativa was regarded as having a simple autopolyploid origin from diploid ssp. caerulea, whereas the autopolyploid origin of tetraploid ssp. falcata from diploid form ssp. falcata is still in doubt. In this study, detailed comparative cytogenetic analysis between diploid to tetraploid species, as well as genomic affinity across different species in the M. sativa complex, were conducted based on comparative mapping of 11 repeated DNA sequences and two rDNA sequences by a fluorescence in situ hybridization (FISH) technique.
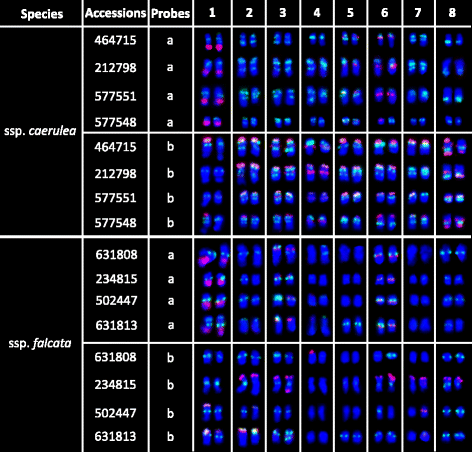
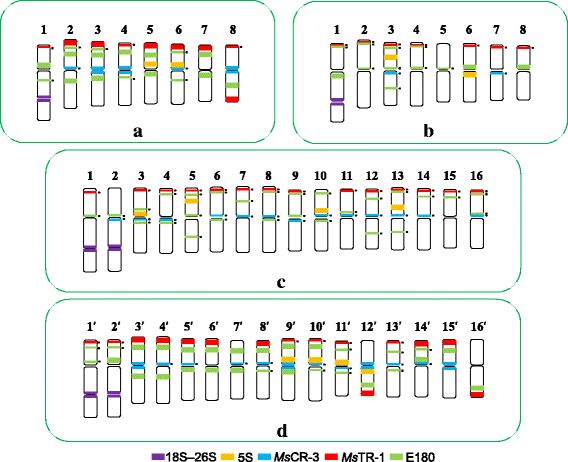
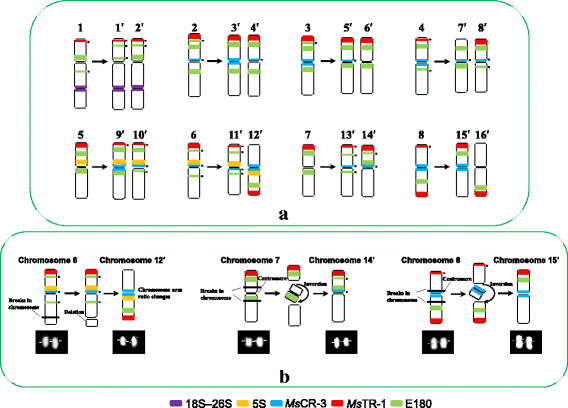
Results: FISH patterns of the repeats in diploid subspecies caerulea were highly similar to those in tetraploid subspecies sativa. Distinctly different FISH patterns were first observed in diploid ssp. falcata, with only centromeric hybridizations using centromeric and multiple region repeats and a few subtelomeric hybridizations using subtelomeric repeats. Tetraploid subspecies falcata was unexpectedly found to possess a highly variable karyotype, which agreed with neither diploid ssp. falcata nor ssp. sativa. Reconstruction of chromosome-doubling process of diploid ssp. caerulea showed that chromosome changes have occurred during polyploidization process.
Conclusions: The comparative cytogenetic results provide reliable evidence that diploid subspecies caerulea is the direct progenitor of tetraploid subspecies sativa. And autotetraploid ssp. sativa has been suggested to undergo a partial diploidization by the progressive accumulation of chromosome structural rearrangements during evolution. However, the tetraploid subspecies falcata is far from a simple autopolyploid from diploid subspecies falcata although no obvious morphological change was observed between these two subspecies.
Keywords: Chromosome evolution; Diploidization; FISH; M. sativa ssp. caerulea; M. sativa ssp. falcata; Medicago sativa; Repetitive sequences.
Figures



Similar articles
-
Karyotypic analysis of N-banded chromosomes of diploid alfalfa: Medicago sativa ssp. caerulea and ssp. falcata and their hybrid.J Hered. 1998 Mar-Apr;89(2):191-3. doi: 10.1093/jhered/89.2.191. J Hered. 1998. PMID: 9542165
-
Karyotypic analysis of C-banded chromosomes of diploid alfalfa: Medicago sativa ssp. caerulea and ssp. falcata and their hybrid.J Hered. 1997 Nov-Dec;88(6):533-7. doi: 10.1093/oxfordjournals.jhered.a023152. J Hered. 1997. PMID: 9419896
-
Physical mapping of rRNA genes in Medicago sativa and M. glomerata by fluorescent in situ hybridization.J Hered. 2000 May-Jun;91(3):256-60. doi: 10.1093/jhered/91.3.256. J Hered. 2000. PMID: 10833055
-
Genetic regulation of meiosis in polyploid species: new insights into an old question.New Phytol. 2010 Apr;186(1):29-36. doi: 10.1111/j.1469-8137.2009.03084.x. Epub 2009 Nov 11. New Phytol. 2010. PMID: 19912546 Review.
-
Towards an understanding of the molecular mechanisms regulating gene expression during diploidization in phylogenetically polyploid lower vertebrates.Hum Genet. 1983;65(1):11-8. doi: 10.1007/BF00285022. Hum Genet. 1983. PMID: 6357994 Review.
Cited by
-
Oligo-FISH barcode chromosome identification system provides novel insights into the natural chromosome aberrations propensity in the autotetraploid cultivated alfalfa.Hortic Res. 2024 Sep 20;12(1):uhae266. doi: 10.1093/hr/uhae266. eCollection 2025 Jan. Hortic Res. 2024. PMID: 39802739 Free PMC article.
-
A haplotype-resolved genome assembly of tetraploid Medicago sativa ssp. falcata.Sci China Life Sci. 2025 Apr;68(4):1186-1189. doi: 10.1007/s11427-024-2753-2. Epub 2024 Dec 20. Sci China Life Sci. 2025. PMID: 39724396 No abstract available.
-
The chromosome-level assembly of the wild diploid alfalfa genome provides insights into the full landscape of genomic variations between cultivated and wild alfalfa.Plant Biotechnol J. 2024 Jun;22(6):1757-1772. doi: 10.1111/pbi.14300. Epub 2024 Jan 30. Plant Biotechnol J. 2024. PMID: 38288521 Free PMC article.
-
Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms.BMC Plant Biol. 2018 Feb 15;18(1):35. doi: 10.1186/s12870-018-1250-4. BMC Plant Biol. 2018. PMID: 29448940 Free PMC article.
-
Low cytomolecular diversification in the genus Stylosanthes Sw. (Papilionoideae, Leguminosae).Genet Mol Biol. 2020 Mar 6;43(1):e20180250. doi: 10.1590/1678-4685-GMB-2018-0250. eCollection 2020. Genet Mol Biol. 2020. PMID: 31429856 Free PMC article.
References
-
- Lim KY, Matyášek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc. 2004;82(4):599–606. doi: 10.1111/j.1095-8312.2004.00344.x. - DOI
-
- Chester M, Gallagher JP, Symonds VV, da Silva AVC, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci U S A. 2012;109(4):1176–1181. doi: 10.1073/pnas.1112041109. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

