New insights into the tetraspanin Tspan5 using novel monoclonal antibodies
- PMID: 28428248
- PMCID: PMC5465482
- DOI: 10.1074/jbc.M116.765669
New insights into the tetraspanin Tspan5 using novel monoclonal antibodies
Abstract
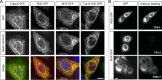
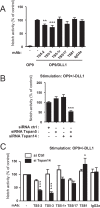
Tspan5 is a member of a subgroup of tetraspanins referred to as TspanC8. These tetraspanins directly interact with the metalloprotease ADAM10, regulate its exit from the endoplasmic reticulum and subsequent trafficking, and differentially regulate its ability to cleave various substrates and activate Notch signaling. The study of Tspan5 has been limited by the lack of good antibodies. This study provides new insights into Tspan5 using new monoclonal antibodies (mAbs), including two mAbs recognizing both Tspan5 and the highly similar tetraspanin Tspan17. Using these mAbs, we show that endogenous Tspan5 associates with ADAM10 in human cell lines and in mouse tissues where it is the most abundant, such as the brain, the lung, the kidney, or the intestine. We also uncover two TspanC8-specific motifs in the large extracellular domain of Tspan5 that are important for ADAM10 interaction and exit from the endoplasmic reticulum. One of the anti-Tspan5 mAbs does not recognize Tspan5 associated with ADAM10, providing a convenient way to measure the fraction of Tspan5 not associated with ADAM10. This fraction is minor in the cell lines tested, and it increases upon transfection of cells with TspanC8 tetraspanins such as Tspan15 or Tspan33 that inhibit Notch signaling. Finally, two antibodies inhibit ligand-induced Notch signaling, and this effect is stronger in cells depleted of the TspanC8 tetraspanin Tspan14, further indicating that Tspan5 and Tspan14 can compensate for each other in Notch signaling.
Keywords: ADAM; ADAM10; Notch pathway; Tspan5; intracellular trafficking; metalloprotease; monoclonal antibody; tetraspanin.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins.Biochem Soc Trans. 2017 Aug 15;45(4):937-44. doi: 10.1042/BST20160296. Epub 2017 Jul 7. Biochem Soc Trans. 2017. PMID: 28687716 Review.
-
TspanC8 tetraspanins differentially regulate ADAM10 endocytosis and half-life.Life Sci Alliance. 2019 Dec 2;3(1):e201900444. doi: 10.26508/lsa.201900444. Print 2020 Jan. Life Sci Alliance. 2019. PMID: 31792032 Free PMC article.
-
TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals.J Cell Biol. 2012 Oct 29;199(3):481-96. doi: 10.1083/jcb.201201133. Epub 2012 Oct 22. J Cell Biol. 2012. PMID: 23091066 Free PMC article.
-
TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization.Cell Mol Life Sci. 2016 May;73(9):1895-915. doi: 10.1007/s00018-015-2111-z. Epub 2015 Dec 19. Cell Mol Life Sci. 2016. PMID: 26686862 Free PMC article.
-
Regulation of ADAM10 by the TspanC8 Family of Tetraspanins and Their Therapeutic Potential.Int J Mol Sci. 2021 Jun 23;22(13):6707. doi: 10.3390/ijms22136707. Int J Mol Sci. 2021. PMID: 34201472 Free PMC article. Review.
Cited by
-
MiR-322-5p Alleviates Cell Injury and Impairment of Cognitive Function in Vascular Dementia by Targeting TSPAN5.Yonsei Med J. 2022 Mar;63(3):282-291. doi: 10.3349/ymj.2022.63.3.282. Yonsei Med J. 2022. PMID: 35184431 Free PMC article.
-
Tetraspanin 5 (TSPAN5), a Novel Gatekeeper of the Tumor Suppressor DLC1 and Myocardin-Related Transcription Factors (MRTFs), Controls HCC Growth and Senescence.Cancers (Basel). 2021 Oct 26;13(21):5373. doi: 10.3390/cancers13215373. Cancers (Basel). 2021. PMID: 34771537 Free PMC article.
-
Calcium-sensing receptor- and ADAM10-mediated klotho shedding is regulated by tetraspanin 5.FEBS Lett. 2025 Mar;599(6):866-875. doi: 10.1002/1873-3468.15078. Epub 2025 Jan 7. FEBS Lett. 2025. PMID: 39777735 Free PMC article.
-
The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex.J Biol Chem. 2020 Sep 4;295(36):12822-12839. doi: 10.1074/jbc.RA120.012601. Epub 2020 Feb 28. J Biol Chem. 2020. PMID: 32111735 Free PMC article.
-
Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins.J Extracell Vesicles. 2020 Apr 16;9(1):1750810. doi: 10.1080/20013078.2020.1750810. eCollection 2020. J Extracell Vesicles. 2020. PMID: 32363014 Free PMC article.
References
-
- Charrin S., Jouannet S., Boucheix C., and Rubinstein E. (2014) Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 - PubMed
-
- Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., and Rubinstein E. (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 - PubMed
-
- Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 - PubMed
-
- Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., and Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 - PubMed
-
- Serru V., Dessen P., Boucheix C., and Rubinstein E. (2000) Sequence and expression of seven new tetraspans. Biochim. Biophys. Acta 1478, 159–163 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

