Crystal structure and redox properties of a novel cyanobacterial heme protein with a His/Cys heme axial ligation and a Per-Arnt-Sim (PAS)-like domain
- PMID: 28428249
- PMCID: PMC5465485
- DOI: 10.1074/jbc.M116.746263
Crystal structure and redox properties of a novel cyanobacterial heme protein with a His/Cys heme axial ligation and a Per-Arnt-Sim (PAS)-like domain
Abstract
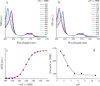
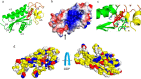
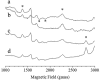
Photosystem II catalyzes light-induced water oxidation leading to the generation of dioxygen indispensable for sustaining aerobic life on Earth. The Photosystem II reaction center is composed of D1 and D2 proteins encoded by psbA and psbD genes, respectively. In cyanobacteria, different psbA genes are present in the genome. The thermophilic cyanobacterium Thermosynechococcus elongatus contains three psbA genes: psbA1, psbA2, and psbA3, and a new c-type heme protein, Tll0287, was found to be expressed in a strain expressing the psbA2 gene only, but the structure and function of Tll0287 are unknown. Here we solved the crystal structure of Tll0287 at a 2.0 Å resolution. The overall structure of Tll0287 was found to be similar to some kinases and sensor proteins with a Per-Arnt-Sim-like domain rather than to other c-type cytochromes. The fifth and sixth axial ligands for the heme were Cys and His, instead of the His/Met or His/His ligand pairs observed for most of the c-type hemes. The redox potential, E½, of Tll0287 was -255 ± 20 mV versus normal hydrogen electrode at pH values above 7.5. Below this pH value, the E½ increased by ≈57 mV/pH unit at 15 °C, suggesting the involvement of a protonatable group with a pKred = 7.2 ± 0.3. Possible functions of Tll0287 as a redox sensor under microaerobic conditions or a cytochrome subunit of an H2S-oxidizing system are discussed in view of the environmental conditions in which psbA2 is expressed, as well as phylogenetic analysis, structural, and sequence homologies.
Keywords: D1 protein; His-Cys heme axial coordination; PAS domain; PAS-like domain; Tll0287; X-ray crystallography; cytochrome; heme; photosynthesis; photosystem II.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


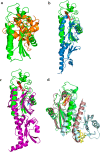


References
-
- Umena Y., Kawakami K., Shen J.-R., and Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 - PubMed
-
- Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., Shimizu T., Yamashita K., Yamamoto M., Ago H., and Shen J.-R. (2015) Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 - PubMed
-
- Wydrzynski T. J., and Satoh K. (eds.) (2005) Photosystem II, the Light-Driven Water: Plastoquinone Oxidoreductase, Springer, Dordrecht, The Netherlands
-
- Shen J.-R. (2015) Structure of Photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 - PubMed
-
- Kawakami K., Umena Y., Kamiya N., and Shen J.-R. (2011) Structure of the catalytic, inorganic core of oxygen-evolving Photosystem II at 1.9 Å resolution. J. Photochem. Photobiol. B 104, 9–18 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

