Poly-protein G-expressing bacteria enhance the sensitivity of immunoassays
- PMID: 28428542
- PMCID: PMC5430508
- DOI: 10.1038/s41598-017-01022-w
Poly-protein G-expressing bacteria enhance the sensitivity of immunoassays
Erratum in
-
Author Correction: Poly-protein G-expressing bacteria enhance the sensitivity of immunoassays.Sci Rep. 2018 Mar 6;8(1):4256. doi: 10.1038/s41598-018-22117-y. Sci Rep. 2018. PMID: 29511251 Free PMC article.
Abstract
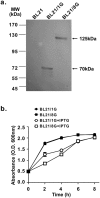
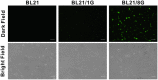
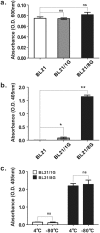
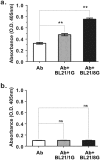
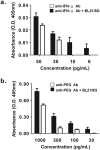
The sensitivities of solid-phase immunoassays are limited by the quantity of detection antibodies bound to their antigens on the solid phase. Here, we developed a poly-protein G-expressing bacterium as an antibody-trapping microparticle to enhance the signals of immunoassays by increasing the accumulation of detection antibodies on the given antigen. Eight tandemly repeated fragment crystallisable (Fc) binding domains of protein G were stably expressed on the surface of Escherichia coli BL21 cells (termed BL21/8G). BL21/8G cells showed a higher avidity for trapping antibodies on their surface than monomeric protein G-expressing BL21 (BL21/1G) cells did. In the sandwich enzyme-linked immunosorbent assay (ELISA), simply mixing the detection antibody with BL21/8G provided a detection limit of 6 pg/mL for human interferon-α (IFN-α) and a limit of 30 pg/mL for polyethylene glycol (PEG)-conjugated IFN-α (Pegasys), which are better than that of the traditional ELISA (30 pg/mL for IFN-α and 100 pg/mL for Pegasys). Moreover, the sensitivity of the Western blot for low-abundance Pegasys (0.4 ng/well) was increased by 25 folds upon mixing of an anti-PEG antibody with BL21/8G cells. By simply being mixed with a detection antibody, the poly-protein G-expressing bacteria can provide a new method to sensitively detect low-abundance target molecules in solid-phase immunoassays.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Kragstrup TW, Vorup-Jensen T, Deleuran B, Hvid M. A simple set of validation steps identifies and removes false results in a sandwich enzyme-linked immunosorbent assay caused by anti-animal IgG antibodies in plasma from arthritis patients. Springerplus. 2013;2:263–270. doi: 10.1186/2193-1801-2-263. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

