Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae
- PMID: 28428552
- PMCID: PMC5430533
- DOI: 10.1038/s41598-017-01144-1
Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae
Abstract
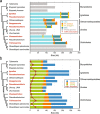
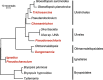
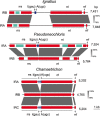
The chloroplast genomes of many algae and almost all land plants carry two identical copies of a large inverted repeat (IR) sequence that can pair for flip-flop recombination and undergo expansion/contraction. Although the IR has been lost multiple times during the evolution of the green algae, the underlying mechanisms are still largely unknown. A recent comparison of IR-lacking and IR-containing chloroplast genomes of chlorophytes from the Ulvophyceae (Ulotrichales) suggested that differential elimination of genes from the IR copies might lead to IR loss. To gain deeper insights into the evolutionary history of the chloroplast genome in the Ulvophyceae, we analyzed the genomes of Ignatius tetrasporus and Pseudocharacium americanum (Ignatiales, an order not previously sampled), Dangemannia microcystis (Oltmannsiellopsidales), Pseudoneochloris marina (Ulvales) and also Chamaetrichon capsulatum and Trichosarcina mucosa (Ulotrichales). Our comparison of these six chloroplast genomes with those previously reported for nine ulvophyceans revealed unsuspected variability. All newly examined genomes feature an IR, but remarkably, the copies of the IR present in the Ignatiales, Pseudoneochloris, and Chamaetrichon diverge in sequence, with the tRNA genes from the rRNA operon missing in one IR copy. The implications of this unprecedented finding for the mechanism of IR loss and flip-flop recombination are discussed.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Lang, B. F. & Nedelcu, A. M. In Genomics of Chloroplasts and Mitochondria Vol. 35 Advances in Photosynthesis and Respiration (eds Ralph Bock & Volker Knoop) Ch. 3, 59–87 (Springer Netherlands, 2012).
-
- Jansen, R. K. & Ruhlman, T. A. In Genomics of Chloroplasts and Mitochondria Vol. 35 Advances in Photosynthesis and Respiration (eds Ralph Bock & Volker Knoop) Ch. 5, 103–126 (Springer Netherlands, 2012).
-
- Brouard JS, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

