Analysis of the Listeria monocytogenes Population Structure among Isolates from 1931 to 2015 in Australia
- PMID: 28428781
- PMCID: PMC5382192
- DOI: 10.3389/fmicb.2017.00603
Analysis of the Listeria monocytogenes Population Structure among Isolates from 1931 to 2015 in Australia
Abstract
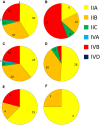
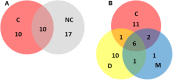
Listeriosis remains among the most important bacterial illnesses, with a high associated mortality rate. Efforts to control listeriosis require detailed knowledge of the epidemiology of the disease itself, and its etiological bacterium, Listeria monocytogenes. In this study we provide an in-depth analysis of the epidemiology of 224 L. monocytogenes isolates from Australian clinical and non-clinical sources. Non-human sources included meat, dairy, seafood, fruit, and vegetables, along with animal and environmental isolates. Serotyping, Multi-Locus Sequence Typing, and analysis of inlA gene sequence were performed. Serogroups IIA, IIB, and IVB comprised 94% of all isolates, with IVB over-represented among clinical isolates. Serogroup IIA was the most common among dairy and meat isolates. Lineage I isolates were most common among clinical isolates, and 52% of clinical isolates belonged to ST1. Overall 39 STs were identified in this study, with ST1 and ST3 containing the largest numbers of L. monocytogenes isolates. These STs comprised 40% of the total isolates (n = 90), and both harbored isolates from clinical and non-clinical sources. ST204 was the third most common ST. The high prevalence of this group among L. monocytogenes populations has not been reported outside Australia. Twenty-seven percent of the STs in this study contained exclusively clinical isolates. Analysis of the virulence protein InlA among isolates in this study identified a truncated form of the protein among isolates from ST121 and ST325. The ST325 group contained a previously unreported novel mutation leading to production of a 93 amino acid protein. This study provides insights in the population structure of L. monocytogenes isolated in Australia, which will contribute to public health knowledge relating to this important human pathogen.
Keywords: Listeria monocytogenes; MLST; SNP typing; inlA; serotype.
Figures




Similar articles
-
Population Structure of Non-ST6 Listeria monocytogenes Isolated in the Red Meat and Poultry Value Chain in South Africa.Microorganisms. 2020 Jul 30;8(8):1152. doi: 10.3390/microorganisms8081152. Microorganisms. 2020. PMID: 32751410 Free PMC article.
-
Genetic characteristics and virulence of Listeria monocytogenes isolated from fresh vegetables in China.BMC Microbiol. 2019 Jun 3;19(1):119. doi: 10.1186/s12866-019-1488-5. BMC Microbiol. 2019. PMID: 31159734 Free PMC article.
-
The Majority of Genotypes of the Virulence Gene inlA Are Intact among Natural Watershed Isolates of Listeria monocytogenes from the Central California Coast.PLoS One. 2016 Dec 1;11(12):e0167566. doi: 10.1371/journal.pone.0167566. eCollection 2016. PLoS One. 2016. PMID: 27907153 Free PMC article.
-
Analysis of Multilocus Sequence Typing and Virulence Characterization of Listeria monocytogenes Isolates from Chinese Retail Ready-to-Eat Food.Front Microbiol. 2016 Feb 16;7:168. doi: 10.3389/fmicb.2016.00168. eCollection 2016. Front Microbiol. 2016. PMID: 26909076 Free PMC article.
-
Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics.Int J Med Microbiol. 2011 Feb;301(2):79-96. doi: 10.1016/j.ijmm.2010.05.002. Epub 2010 Aug 13. Int J Med Microbiol. 2011. PMID: 20708964 Review.
Cited by
-
Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland.Front Microbiol. 2020 Jun 25;11:1412. doi: 10.3389/fmicb.2020.01412. eCollection 2020. Front Microbiol. 2020. PMID: 32670248 Free PMC article.
-
Isolation and Characterization of Clinical Listeria monocytogenes in Beijing, China, 2014-2016.Front Microbiol. 2019 May 8;10:981. doi: 10.3389/fmicb.2019.00981. eCollection 2019. Front Microbiol. 2019. PMID: 31139159 Free PMC article.
-
Human Listeriosis.Clin Microbiol Rev. 2023 Mar 23;36(1):e0006019. doi: 10.1128/cmr.00060-19. Epub 2022 Dec 8. Clin Microbiol Rev. 2023. PMID: 36475874 Free PMC article. Review.
-
Phylogenetically Defined Isoforms of Listeria monocytogenes Invasion Factor InlB Differently Activate Intracellular Signaling Pathways and Interact with the Receptor gC1q-R.Int J Mol Sci. 2019 Aug 24;20(17):4138. doi: 10.3390/ijms20174138. Int J Mol Sci. 2019. PMID: 31450632 Free PMC article.
-
Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains.Genes (Basel). 2018 Feb 9;9(2):80. doi: 10.3390/genes9020080. Genes (Basel). 2018. PMID: 29425131 Free PMC article.
References
-
- Bille J., Rocourt J. (1996). WHO international multicenter Listeria monocytogenes subtyping study - rationale and set-up of the study. Int. J. Food Microbiol. 32 12. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

