Non-canonical Translation in Plant RNA Viruses
- PMID: 28428795
- PMCID: PMC5382211
- DOI: 10.3389/fpls.2017.00494
Non-canonical Translation in Plant RNA Viruses
Abstract
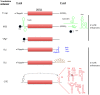
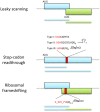
Viral protein synthesis is completely dependent upon the host cell's translational machinery. Canonical translation of host mRNAs depends on structural elements such as the 5' cap structure and/or the 3' poly(A) tail of the mRNAs. Although many viral mRNAs are devoid of one or both of these structures, they can still translate efficiently using non-canonical mechanisms. Here, we review the tools utilized by positive-sense single-stranded (+ss) RNA plant viruses to initiate non-canonical translation, focusing on cis-acting sequences present in viral mRNAs. We highlight how these elements may interact with host translation factors and speculate on their contribution for achieving translational control. We also describe other translation strategies used by plant viruses to optimize the usage of the coding capacity of their very compact genomes, including leaky scanning initiation, ribosomal frameshifting and stop-codon readthrough. Finally, future research perspectives on the unusual translational strategies of +ssRNA viruses are discussed, including parallelisms between viral and host mRNAs mechanisms of translation, particularly for host mRNAs which are translated under stress conditions.
Keywords: 3′-CITE; IRES; RNA structure and function; non-canonical translation; protein synthesis; translation enhancers; translational recoding.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

