Genome-wide search for candidate genes for yeast robustness improvement against formic acid reveals novel susceptibility (Trk1 and positive regulators) and resistance (Haa1-regulon) determinants
- PMID: 28428821
- PMCID: PMC5395885
- DOI: 10.1186/s13068-017-0781-5
Genome-wide search for candidate genes for yeast robustness improvement against formic acid reveals novel susceptibility (Trk1 and positive regulators) and resistance (Haa1-regulon) determinants
Abstract
Background: Formic acid is an inhibitory compound present in lignocellulosic hydrolysates. Understanding the complex molecular mechanisms underlying Saccharomyces cerevisiae tolerance to this weak acid at the system level is instrumental to guide synthetic pathway engineering for robustness improvement of industrial strains envisaging their use in lignocellulosic biorefineries.
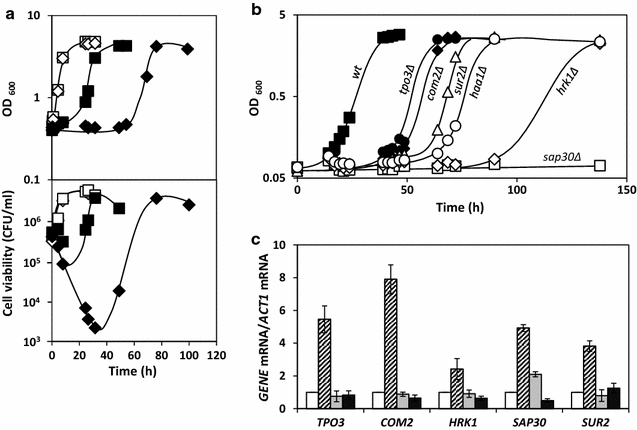
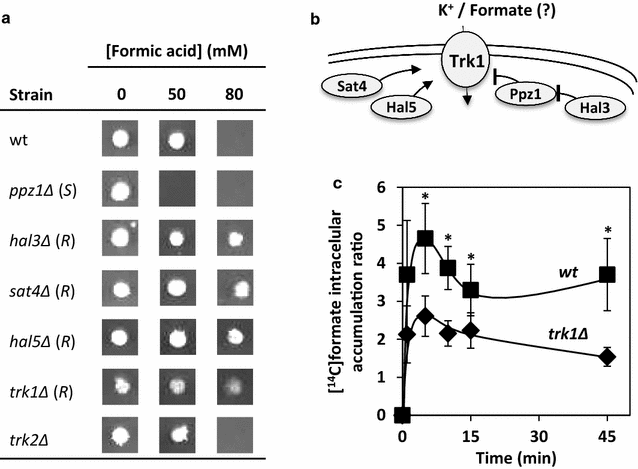
Results: This study was performed to identify, at a genome-wide scale, genes whose expression confers protection or susceptibility to formic acid, based on the screening of a haploid deletion mutant collection to search for these phenotypes in the presence of 60, 70 and 80 mM of this acid, at pH 4.5. This chemogenomic analysis allowed the identification of 172 determinants of tolerance and 41 determinants of susceptibility to formic acid. Clustering of genes required for maximal tolerance to this weak acid, based on their biological function, indicates an enrichment of those involved in intracellular trafficking and protein synthesis, cell wall and cytoskeleton organization, carbohydrate metabolism, lipid, amino acid and vitamin metabolism, response to stress, chromatin remodelling, transcription and internal pH homeostasis. Among these genes is HAA1 encoding the main transcriptional regulator of yeast transcriptome reprograming in response to acetic acid and genes of the Haa1-regulon; all demonstrated determinants of acetic acid tolerance. Among the genes that when deleted lead to increased tolerance to formic acid, TRK1, encoding the high-affinity potassium transporter and a determinant of resistance to acetic acid, was surprisingly found. Consistently, genes encoding positive regulators of Trk1 activity were also identified as formic acid susceptibility determinants, while a negative regulator confers protection. At a saturating K+ concentration of 20 mM, the deletion mutant trk1Δ was found to exhibit a much higher tolerance compared with the parental strain. Given that trk1Δ accumulates lower levels of radiolabelled formic acid, compared to the parental strain, it is hypothesized that Trk1 facilitates formic acid uptake into the yeast cell.
Conclusions: The list of genes resulting from this study shows a few marked differences from the list of genes conferring protection to acetic acid and provides potentially valuable information to guide improvement programmes for the development of more robust strains against formic acid.
Keywords: Chemogenomic analysis; Formic acid tolerance; Formic acid toxicity; Haa1; Lignocellulosic hydrolysates; Trk1; Yeast robustness.
Figures


Similar articles
-
Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid.Microb Cell Fact. 2010 Oct 25;9:79. doi: 10.1186/1475-2859-9-79. Microb Cell Fact. 2010. PMID: 20973990 Free PMC article.
-
Improvement of yeast tolerance to acetic acid through Haa1 transcription factor engineering: towards the underlying mechanisms.Microb Cell Fact. 2017 Jan 9;16(1):7. doi: 10.1186/s12934-016-0621-5. Microb Cell Fact. 2017. PMID: 28068993 Free PMC article.
-
Search for genes responsible for the remarkably high acetic acid tolerance of a Zygosaccharomyces bailii-derived interspecies hybrid strain.BMC Genomics. 2015 Dec 16;16:1070. doi: 10.1186/s12864-015-2278-6. BMC Genomics. 2015. PMID: 26673744 Free PMC article.
-
[Advances in functional genomics studies underlying acetic acid tolerance of Saccharomyces cerevisiae].Sheng Wu Gong Cheng Xue Bao. 2014 Mar;30(3):368-80. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25007573 Review. Chinese.
-
Adaptive Response and Tolerance to Acetic Acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: A Physiological Genomics Perspective.Front Microbiol. 2018 Feb 21;9:274. doi: 10.3389/fmicb.2018.00274. eCollection 2018. Front Microbiol. 2018. PMID: 29515554 Free PMC article. Review.
Cited by
-
Genome-Wide Identification of Genes Involved in General Acid Stress and Fluoride Toxicity in Saccharomyces cerevisiae.Front Microbiol. 2020 Jun 25;11:1410. doi: 10.3389/fmicb.2020.01410. eCollection 2020. Front Microbiol. 2020. PMID: 32670247 Free PMC article.
-
CRISPRi screen highlights chromatin regulation to be involved in formic acid tolerance in Saccharomyces cerevisiae.Eng Microbiol. 2023 Feb 3;3(2):100076. doi: 10.1016/j.engmic.2023.100076. eCollection 2023 Jun. Eng Microbiol. 2023. PMID: 39629247 Free PMC article.
-
The NPR/Hal family of protein kinases in yeasts: biological role, phylogeny and regulation under environmental challenges.Comput Struct Biotechnol J. 2022 Oct 15;20:5698-5712. doi: 10.1016/j.csbj.2022.10.006. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36320937 Free PMC article. Review.
-
Changes in lipid metabolism convey acid tolerance in Saccharomyces cerevisiae.Biotechnol Biofuels. 2018 Oct 29;11:297. doi: 10.1186/s13068-018-1295-5. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30450126 Free PMC article.
-
How adaptive laboratory evolution can boost yeast tolerance to lignocellulosic hydrolyses.Curr Genet. 2022 Aug;68(3-4):319-342. doi: 10.1007/s00294-022-01237-z. Epub 2022 Apr 1. Curr Genet. 2022. PMID: 35362784 Review.
References
-
- Sangster J. Octanol–water partition-coefficients of simple organic-compounds. J Phys Chem Ref Data. 1989;18(3):1111–1229. doi: 10.1063/1.555833. - DOI
-
- Reutemann W, Kieczka H. Formic acid. In: Ullmann’s encyclopedia of industrial chemistry. Wiley-VCH Verlag GmbH & Co. KGaA; 2000.
-
- Greetham D, Wimalasena T, Kerruish DW, Brindley S, Ibbett RN, Linforth RL, Tucker G, Phister TG, Smart KA. Development of a phenotypic assay for characterisation of ethanologenic yeast strain sensitivity to inhibitors released from lignocellulosic feedstocks. J Ind Microbiol Biotechnol. 2014;41(6):931–945. doi: 10.1007/s10295-014-1431-6. - DOI - PubMed
-
- Tomas-Pejo E, Oliva JM, Ballesteros M, Olsson L. Comparison of SHF and SSF processes from steam-exploded wheat straw for ethanol production by xylose-fermenting and robust glucose-fermenting Saccharomyces cerevisiae strains. Biotechnol Bioeng. 2008;100(6):1122–1131. doi: 10.1002/bit.21849. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

