Detection of 1p36 deletion by clinical exome-first diagnostic approach
- PMID: 28428889
- PMCID: PMC5381605
- DOI: 10.1038/hgv.2016.6
Detection of 1p36 deletion by clinical exome-first diagnostic approach
Abstract
Although chromosome 1p36 deletion syndrome is considered clinically recognizable based on characteristic features, the clinical manifestations of patients during infancy are often not consistent with those observed later in life. We report a 4-month-old girl who showed multiple congenital anomalies and developmental delay, but no clinical signs of syndromic disease caused by a terminal deletion in 1p36.32-p36.33 that was first identified by targeted-exome sequencing for molecular diagnosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
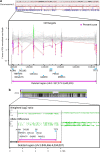
References
-
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 2000; 34: 297–329. - PubMed
-
- Heilstedt HA, Ballif BC, Howard LA, Kashork CD, Shaffer LG. Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Genet 2003; 64: 310–316. - PubMed
-
- Battaglia A. 1p36 deletion syndrome. In:Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A (eds). GeneReviews [Internet]. University of Washington, Seattle: Seattle, WA, 1993–2015.
Data Citations
-
- Imoto Issei.HGV Database. 2016. 10.6084/m9.figshare.hgv.784. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

