Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom
- PMID: 28429538
- PMCID: PMC5485032
- DOI: 10.1111/nph.14557
Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom
Abstract
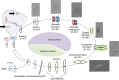
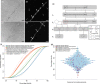
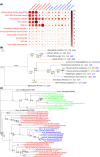
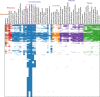
Microalgae play a major role as primary producers in aquatic ecosystems. Cell signalling regulates their interactions with the environment and other organisms, yet this process in phytoplankton is poorly defined. Using the marine planktonic diatom Pseudo-nitzschia multistriata, we investigated the cell response to cues released during sexual reproduction, an event that demands strong regulatory mechanisms and impacts on population dynamics. We sequenced the genome of P. multistriata and performed phylogenomic and transcriptomic analyses, which allowed the definition of gene gains and losses, horizontal gene transfers, conservation and evolutionary rate of sex-related genes. We also identified a small number of conserved noncoding elements. Sexual reproduction impacted on cell cycle progression and induced an asymmetric response of the opposite mating types. G protein-coupled receptors and cyclic guanosine monophosphate (cGMP) are implicated in the response to sexual cues, which overall entails a modulation of cell cycle, meiosis-related and nutrient transporter genes, suggesting a fine control of nutrient uptake even under nutrient-replete conditions. The controllable life cycle and the genome sequence of P. multistriata allow the reconstruction of changes occurring in diatoms in a key phase of their life cycle, providing hints on the evolution and putative function of their genes and empowering studies on sexual reproduction.
Keywords: Pseudo-nitzschia multistriata; algae; diatom; genomics; mating type; phytoplankton; sexual reproduction; signal transduction.
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures

References
-
- Adelfi MG, Borra M, Sanges R, Montresor M, Fontana A, Ferrante MI. 2014. Selection and validation of reference genes for qPCR analysis in the pennate diatoms Pseudo‐nitzschia multistriata and P. arenysensis . Journal of Experimental Marine Biology and Ecology 451: 74–81.
-
- Armbrust EV. 2009. The life of diatoms in the world's oceans. Nature 459: 185–192. - PubMed
-
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M et al 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. - PubMed
-
- Barkai N, Rose MD, Wingreen NS. 1998. Protease helps yeast find mating partners. Nature 396: 422–423. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

