Absolute Binding Free Energies between T4 Lysozyme and 141 Small Molecules: Calculations Based on Multiple Rigid Receptor Configurations
- PMID: 28430432
- PMCID: PMC5612505
- DOI: 10.1021/acs.jctc.6b01183
Absolute Binding Free Energies between T4 Lysozyme and 141 Small Molecules: Calculations Based on Multiple Rigid Receptor Configurations
Abstract
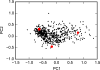
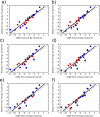
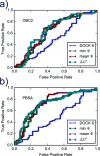
We demonstrate the feasibility of estimating protein-ligand binding free energies using multiple rigid receptor configurations. On the basis of T4 lysozyme snapshots extracted from six alchemical binding free energy calculations with a flexible receptor, binding free energies were estimated for a total of 141 ligands. For 24 ligands, the calculations reproduced flexible-receptor estimates with a correlation coefficient of 0.90 and a root-mean-square error of 1.59 kcal/mol. The accuracy of calculations based on Poisson-Boltzmann/surface area implicit solvent was comparable to that of previously reported free energy calculations.
Figures







Similar articles
-
Using the fast fourier transform in binding free energy calculations.J Comput Chem. 2018 Apr 30;39(11):621-636. doi: 10.1002/jcc.25139. Epub 2017 Dec 22. J Comput Chem. 2018. PMID: 29270990 Free PMC article.
-
Protein-ligand binding free energies from exhaustive docking.J Phys Chem B. 2012 Jun 14;116(23):6872-9. doi: 10.1021/jp212646s. Epub 2012 Apr 2. J Phys Chem B. 2012. PMID: 22432509
-
Nonbonded Force Field Parameters from MBIS Partitioning of the Molecular Electron Density Improve Binding Affinity Predictions of the T4-Lysozyme Double Mutant.J Chem Inf Model. 2024 Apr 22;64(8):3269-3277. doi: 10.1021/acs.jcim.3c01912. Epub 2024 Mar 28. J Chem Inf Model. 2024. PMID: 38546407
-
The tail lysozyme complex of bacteriophage T4.Int J Biochem Cell Biol. 2003 Jan;35(1):16-21. doi: 10.1016/s1357-2725(02)00098-5. Int J Biochem Cell Biol. 2003. PMID: 12467643 Review.
-
Structural and genetic analysis of the folding and function of T4 lysozyme.FASEB J. 1996 Jan;10(1):35-41. doi: 10.1096/fasebj.10.1.8566545. FASEB J. 1996. PMID: 8566545 Review.
Cited by
-
Implicit ligand theory for relative binding free energies: II. An estimator based on control variates.J Phys Commun. 2020 Nov;4(11):115010. doi: 10.1088/2399-6528/abcbac. Epub 2020 Nov 26. J Phys Commun. 2020. PMID: 33817346 Free PMC article.
-
On Restraints in End-Point Protein-Ligand Binding Free Energy Calculations.J Comput Chem. 2020 Mar 5;41(6):573-586. doi: 10.1002/jcc.26119. Epub 2019 Dec 10. J Comput Chem. 2020. PMID: 31821590 Free PMC article.
-
The SAMPL6 SAMPLing challenge: assessing the reliability and efficiency of binding free energy calculations.J Comput Aided Mol Des. 2020 May;34(5):601-633. doi: 10.1007/s10822-020-00290-5. Epub 2020 Jan 27. J Comput Aided Mol Des. 2020. PMID: 31984465 Free PMC article.
-
Alchemical Grid Dock (AlGDock): Binding Free Energy Calculations between Flexible Ligands and Rigid Receptors.J Comput Chem. 2020 Mar 15;41(7):715-730. doi: 10.1002/jcc.26036. Epub 2019 Aug 9. J Comput Chem. 2020. PMID: 31397498 Free PMC article.
-
Temperature artifacts in protein structures bias ligand-binding predictions.Chem Sci. 2021 Jul 13;12(34):11275-11293. doi: 10.1039/d1sc02751d. eCollection 2021 Sep 1. Chem Sci. 2021. PMID: 34667539 Free PMC article.
References
-
- Michel J, Essex JW. Prediction of Protein-Ligand Binding Affinity by Free Energy Simulations: Assumptions, Pitfalls and Expectations. J Comput-Aided Mol Des. 2010;24:639–658. - PubMed
-
- Gilson MK, Zhou H-X. Calculation of Protein-Ligand Binding Affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. - PubMed
-
- Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A Critical Assessment of Docking Programs and Scoring Functions. J Med Chem. 2006;49:5912–5931. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
