Synthetic lethal interaction between the tumour suppressor STAG2 and its paralog STAG1
- PMID: 28430577
- PMCID: PMC5514935
- DOI: 10.18632/oncotarget.16838
Synthetic lethal interaction between the tumour suppressor STAG2 and its paralog STAG1
Abstract
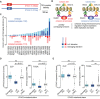
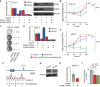
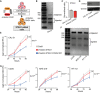
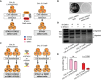
Cohesin is a multi-protein complex that tethers sister chromatids during mitosis and mediates DNA repair, genome compartmentalisation and regulation of gene expression. Cohesin subunits frequently acquire cancer loss-of-function alterations and act as tumour suppressors in several tumour types. This has led to increased interest in cohesin as potential target in anti-cancer therapy. Here we show that the loss-of-function of STAG2, a core component of cohesin and an emerging tumour suppressor, leads to synthetic dependency of mutated cancer cells on its paralog STAG1. STAG1 and STAG2 share high sequence identity, encode mutually exclusive cohesin subunits and retain partially overlapping functions. We inhibited STAG1 and STAG2 in several cancer cell lines where the two genes have variable mutation and copy number status. In all cases, we observed that the simultaneous blocking of STAG1 and STAG2 significantly reduces cell proliferation. We further confirmed the synthetic lethal interaction developing a vector-free CRISPR system to induce STAG1/STAG2 double gene knockout. We provide strong evidence that STAG1 is a promising therapeutic target in cancers with inactivating alterations of STAG2.
Keywords: cancer vulnerability; cohesin complex; paralog dependency; precision medicine; synthetic lethality.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. - PubMed
-
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. - PubMed
-
- Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14:389–393. - PubMed
-
- Mehta GD, Kumar R, Srivastava S, Ghosh SK. Cohesin: functions beyond sister chromatid cohesion. FEBS Lett. 2013;587:2299–2312. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

