Histone demethylase JMJD3 regulates CD11a expression through changes in histone H3K27 tri-methylation levels in CD4+ T cells of patients with systemic lupus erythematosus
- PMID: 28430662
- PMCID: PMC5564738
- DOI: 10.18632/oncotarget.16894
Histone demethylase JMJD3 regulates CD11a expression through changes in histone H3K27 tri-methylation levels in CD4+ T cells of patients with systemic lupus erythematosus
Abstract
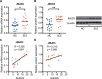
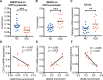
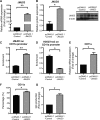
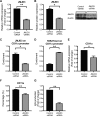
Aberrant CD11a overexpression in CD4+ T cells induces T cell auto-reactivity, which is an important factor for systemic lupus erythematosus (SLE) pathogenesis. Although many studies have focused on CD11a epigenetic regulation, little is known about histone methylation. JMJD3, as a histone demethylase, is capable of specifically removing the trimethyl group from the H3K27 lysine residue, triggering target gene activation. Here, we examined the expression and function of JMJD3 in CD4+ T cells from SLE patients. Significantly decreased H3K27me3 levels and increased JMJD3 binding were detected within the ITGAL (CD11a) promoter locus in SLE CD4+ T cells compared with those in healthy CD4+ T cells. Moreover, overexpressing JMJD3 through the transfection of pcDNA3.1-JMJD3 into healthy donor CD4+ T cells increased JMJD3 enrichment and decreased H3K27me3 enrichment within the ITGAL (CD11a) promoter and up-regulated CD11a expression, leading to T and B cell hyperactivity. Inhibition of JMJD3 via JMJD3-siRNA in SLE CD4+ T cells showed the opposite effects. These results demonstrated that histone demethylase JMJD3 regulates CD11a expression in lupus T cells by affecting the H3K27me3 levels in the ITGAL (CD11a) promoter region, and JMJD3 might thereby serve as a potential therapeutic target for SLE.
Keywords: CD11a; CD4+ T cells; H3K27me3; JMJD3; SLE.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
Similar articles
-
Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus.J Autoimmun. 2011 Nov;37(3):180-9. doi: 10.1016/j.jaut.2011.09.006. Epub 2011 Oct 19. J Autoimmun. 2011. PMID: 22014533
-
RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1.Arthritis Res Ther. 2010;12(6):R227. doi: 10.1186/ar3214. Epub 2010 Dec 30. Arthritis Res Ther. 2010. PMID: 21192791 Free PMC article.
-
[Effect of aberrant H3K27me3 modification in promoter regions on cAMP response element modulator α expression in CD4+ T cells from patients with systemic lupus erythematosus].Nan Fang Yi Ke Da Xue Xue Bao. 2017 Dec 20;37(12):1597-1602. doi: 10.3969/j.issn.1673-4254.2017.12.06. Nan Fang Yi Ke Da Xue Xue Bao. 2017. PMID: 29292251 Free PMC article. Chinese.
-
Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus.J Autoimmun. 2013 Mar;41:92-9. doi: 10.1016/j.jaut.2013.01.005. Epub 2013 Jan 20. J Autoimmun. 2013. PMID: 23340289 Review.
-
Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process.J Mol Med (Berl). 2014 Oct;92(10):1035-43. doi: 10.1007/s00109-014-1182-x. Epub 2014 Jun 14. J Mol Med (Berl). 2014. PMID: 24925089 Review.
Cited by
-
Epigenetic Dysregulation in Autoimmune and Inflammatory Skin Diseases.Clin Rev Allergy Immunol. 2022 Dec;63(3):447-471. doi: 10.1007/s12016-022-08956-8. Epub 2022 Nov 8. Clin Rev Allergy Immunol. 2022. PMID: 36346551 Review.
-
Transcriptional regulation of Tfh dynamics and the formation of immunological synapses.Exp Mol Med. 2024 Jun;56(6):1365-1372. doi: 10.1038/s12276-024-01254-7. Epub 2024 Jun 3. Exp Mol Med. 2024. PMID: 38825646 Free PMC article. Review.
-
Epigenetics in SLE.Curr Rheumatol Rep. 2017 Sep;19(9):58. doi: 10.1007/s11926-017-0685-1. Curr Rheumatol Rep. 2017. PMID: 28752494 Free PMC article. Review.
-
Effects of Major Epigenetic Factors on Systemic Lupus Erythematosus.Iran Biomed J. 2018 Sep;22(5):294-302. doi: 10.29252/ibj.22.5.294. Epub 2018 May 27. Iran Biomed J. 2018. PMID: 29803202 Free PMC article. Review.
-
Immunity and autoantibodies of a mouse strain with autistic-like behavior.Brain Behav Immun Health. 2020 Apr 13;4:100069. doi: 10.1016/j.bbih.2020.100069. eCollection 2020 Apr. Brain Behav Immun Health. 2020. PMID: 34589851 Free PMC article.
References
-
- Invernizzi P. Future directions in genetic for autoimmune diseases. J Autoimmun. 2009;33:1–2. - PubMed
-
- Youinou P, Pers JO, Gershwin ME, Shoenfeld Y. Geo-epidemiology and autoimmunity. J Autoimmun. 2010;34:J163–7. - PubMed
-
- Mohan C, Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat Rev Nephrol. 2015;11:329–41. - PubMed
-
- Teruel M, Alarcon-Riquelme ME. Genetics of systemic lupus erythematosus and Sjogren's syndrome: an update. Curr Opin Rheumatol. 2016;28:506–14. - PubMed
-
- Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

