Estimating optimal sparseness of developmental gene networks using a semi-quantitative model
- PMID: 28430819
- PMCID: PMC5400252
- DOI: 10.1371/journal.pone.0176492
Estimating optimal sparseness of developmental gene networks using a semi-quantitative model
Abstract
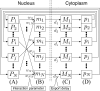
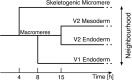
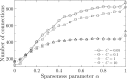
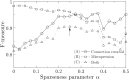
To estimate gene regulatory networks, it is important that we know the number of connections, or sparseness of the networks. It can be expected that the robustness to perturbations is one of the factors determining the sparseness. We reconstruct a semi-quantitative model of gene networks from gene expression data in embryonic development and detect the optimal sparseness against perturbations. The dense networks are robust to connection-removal perturbation, whereas the sparse networks are robust to misexpression perturbation. We show that there is an optimal sparseness that serves as a trade-off between these perturbations, in agreement with the optimal result of validation for testing data. These results suggest that the robustness to the two types of perturbations determines the sparseness of gene networks.
Conflict of interest statement
Figures




Similar articles
-
On the role of sparseness in the evolution of modularity in gene regulatory networks.PLoS Comput Biol. 2018 May 18;14(5):e1006172. doi: 10.1371/journal.pcbi.1006172. eCollection 2018 May. PLoS Comput Biol. 2018. PMID: 29775459 Free PMC article.
-
Asymmetry in indegree and outdegree distributions of gene regulatory networks arising from dynamical robustness.Phys Rev E. 2018 Jun;97(6-1):062315. doi: 10.1103/PhysRevE.97.062315. Phys Rev E. 2018. PMID: 30011527
-
Landscape of gene networks for random parameter perturbation.Integr Biol (Camb). 2018 Feb 19;10(2):92-99. doi: 10.1039/c7ib00198c. Integr Biol (Camb). 2018. PMID: 29340399
-
Reverse engineering gene regulatory networks by modular response analysis - a benchmark.Essays Biochem. 2018 Oct 26;62(4):535-547. doi: 10.1042/EBC20180012. Print 2018 Oct 26. Essays Biochem. 2018. PMID: 30315094 Review.
-
Organization principles in genetic interaction networks.Adv Exp Med Biol. 2012;751:53-78. doi: 10.1007/978-1-4614-3567-9_3. Adv Exp Med Biol. 2012. PMID: 22821453 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

