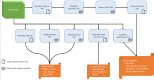
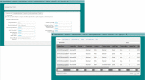
MetaLIMS, a simple open-source laboratory information management system for small metagenomic labs
- PMID: 28430964
- PMCID: PMC5449644
- DOI: 10.1093/gigascience/gix025
MetaLIMS, a simple open-source laboratory information management system for small metagenomic labs
Abstract
As the cost of sequencing continues to fall, smaller groups increasingly initiate and manage larger sequencing projects and take on the complexity of data storage for high volumes of samples. This has created a need for low-cost laboratory information management systems (LIMS) that contain flexible fields to accommodate the unique nature of individual labs. Many labs do not have a dedicated information technology position, so LIMS must also be easy to setup and maintain with minimal technical proficiency. MetaLIMS is a free and open-source web-based application available via GitHub. The focus of MetaLIMS is to store sample metadata prior to sequencing and analysis pipelines. Initially designed for environmental metagenomics labs, in addition to storing generic sample collection information and DNA/RNA processing information, the user can also add fields specific to the user's lab. MetaLIMS can also produce a basic sequencing submission form compatible with the proprietary Clarity LIMS system used by some sequencing facilities. To help ease the technical burden associated with web deployment, MetaLIMS options the use of commercial web hosting combined with MetaLIMS bash scripts for ease of setup. MetaLIMS overcomes key challenges common in LIMS by giving labs access to a low-cost and open-source tool that also has the flexibility to meet individual lab needs and an option for easy deployment. By making the web application open source and hosting it on GitHub, we hope to encourage the community to build upon MetaLIMS, making it more robust and tailored to the needs of more researchers.
Keywords: GitHub; HTML; JavaScript; LIMS; customizable; mysql; open-source; php; sample management; web application.
© The Authors 2017. Published by Oxford University Press.
Figures
Similar articles
-
Development of an open source laboratory information management system for 2-D gel electrophoresis-based proteomics workflow.BMC Bioinformatics. 2006 Oct 4;7:430. doi: 10.1186/1471-2105-7-430. BMC Bioinformatics. 2006. PMID: 17018156 Free PMC article.
-
MASTR-MS: a web-based collaborative laboratory information management system (LIMS) for metabolomics.Metabolomics. 2017;13(2):14. doi: 10.1007/s11306-016-1142-2. Epub 2016 Dec 27. Metabolomics. 2017. PMID: 28090199 Free PMC article.
-
Screensaver: an open source lab information management system (LIMS) for high throughput screening facilities.BMC Bioinformatics. 2010 May 18;11:260. doi: 10.1186/1471-2105-11-260. BMC Bioinformatics. 2010. PMID: 20482787 Free PMC article.
-
Advancing Commercialization of Digital Products from Federal Laboratories.Washington (DC): National Academies Press (US); 2021 Jan 1. Washington (DC): National Academies Press (US); 2021 Jan 1. PMID: 33687821 Free Books & Documents. Review.
-
POC Testing and Reporting of Sodium, and Other Small Molecules Need Modified IFCC Source/Type Designations to Improve Operational Efficacy and for Clinically Accurate, Unambiguous Reporting from LIMS and HIS.EJIFCC. 2023 Dec 21;34(4):271-275. eCollection 2023 Dec. EJIFCC. 2023. PMID: 38303750 Free PMC article. Review. No abstract available.
Cited by
-
LabxDB: versatile databases for genomic sequencing and lab management.Bioinformatics. 2020 Aug 15;36(16):4530-4531. doi: 10.1093/bioinformatics/btaa557. Bioinformatics. 2020. PMID: 32502232 Free PMC article.
-
IOCBIO Kinetics: An open-source software solution for analysis of data traces.PLoS Comput Biol. 2020 Dec 22;16(12):e1008475. doi: 10.1371/journal.pcbi.1008475. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33351800 Free PMC article.
References
-
- Genologics Clarity LIMS www.genologics.com/claritylims (15 February 2016, date last accessed).
-
- Starlims www.abbottinformatics.com/us/products/lims (15 February 2016, date last accessed).
-
- Scholtalbers J, Rossler J, Sorn P et al. . Galaxy LIMS for next-generation sequencing. Bioinformatics 2013; 29:1233–4. - PubMed
-
- SciGenom Labs www.scigenom.com (15 February 2016, date last accessed).
-
- MISO Managing Information for Sequencing Operations http://www.earlham.ac.uk/miso/ (9 November 2016, date last accessed).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous