Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA
- PMID: 28430982
- PMCID: PMC6075211
- DOI: 10.1093/hmg/ddx142
Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA
Erratum in
-
Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA.Hum Mol Genet. 2017 Jul 1;26(13):2590. doi: 10.1093/hmg/ddx220. Hum Mol Genet. 2017. PMID: 28637317 Free PMC article. No abstract available.
Abstract
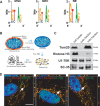
Mitochondria are thought to have originated as free-living prokaryotes. Mitochondria organelles have small circular genomes with substantial structural and genetic similarity to bacteria. Contrary to the prevailing concept of intronless mitochondria, here we present evidence that mitochondrial RNA transcripts (mtRNA) are not limited to policystronic molecules, but also processed as nuclei-like transcripts that are differentially spliced and expressed in a cell-type specific manner. The presence of canonical splice sites in the mtRNA introns and of core components of the nuclei-encoded spliceosome machinery within the mitochondrial organelle suggest that nuclei-encoded spliceosome can mediate splicing of mtRNA.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Asin-Cayuela J., Gustafsson C.M. (2007) Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci., 32, 111–117. - PubMed
-
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet., 5, 123–135. - PubMed
-
- Gray M.W. (1999) Mitochondrial Evolution. Science, 283, 1476–1481. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

