Cis-Regulatory Divergence in Gene Expression between Two Thermally Divergent Yeast Species
- PMID: 28431042
- PMCID: PMC5554586
- DOI: 10.1093/gbe/evx072
Cis-Regulatory Divergence in Gene Expression between Two Thermally Divergent Yeast Species
Abstract
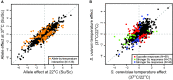
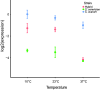
Gene regulation is a ubiquitous mechanism by which organisms respond to their environment. While organisms are often found to be adapted to the environments they experience, the role of gene regulation in environmental adaptation is not often known. In this study, we examine divergence in cis-regulatory effects between two Saccharomycesspecies, S. cerevisiaeand S. uvarum, that have substantially diverged in their thermal growth profile. We measured allele specific expression (ASE) in the species' hybrid at three temperatures, the highest of which is lethal to S. uvarumbut not the hybrid or S. cerevisiae. We find that S. uvarumalleles can be expressed at the same level as S. cerevisiaealleles at high temperature and most cis-acting differences in gene expression are not dependent on temperature. While a small set of 136 genes show temperature-dependent ASE, we find no indication that signatures of directional cis-regulatory evolution are associated with temperature. Within promoter regions we find binding sites enriched upstream of temperature responsive genes, but only weak correlations between binding site and expression divergence. Our results indicate that temperature divergence between S. cerevisiaeand S. uvarumhas not caused widespread divergence in cis-regulatory activity, but point to a small subset of genes where the species' alleles show differences in magnitude or opposite responses to temperature. The difficulty of explaining divergence in cis-regulatory sequences with models of transcription factor binding sites and nucleosome positioning highlights the importance of identifying mutations that underlie cis-regulatory divergence between species.
Keywords: Saccharomyces; allele-specific expression; cis-regulatory evolution; gene expression; interspecific hybrid; thermotolerance.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Evolutionary Dynamics of Regulatory Changes Underlying Gene Expression Divergence among Saccharomyces Species.Genome Biol Evol. 2017 Apr 1;9(4):843-854. doi: 10.1093/gbe/evx035. Genome Biol Evol. 2017. PMID: 28338820 Free PMC article.
-
A yeast hybrid provides insight into the evolution of gene expression regulation.Science. 2009 May 1;324(5927):659-62. doi: 10.1126/science.1169766. Science. 2009. PMID: 19407207
-
Polygenic and directional regulatory evolution across pathways in Saccharomyces.Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):5058-63. doi: 10.1073/pnas.0912959107. Epub 2010 Mar 1. Proc Natl Acad Sci U S A. 2010. PMID: 20194736 Free PMC article.
-
Tempo and mode in evolution of transcriptional regulation.PLoS Genet. 2012 Jan;8(1):e1002432. doi: 10.1371/journal.pgen.1002432. Epub 2012 Jan 19. PLoS Genet. 2012. PMID: 22291600 Free PMC article. Review.
-
Fungal regulatory evolution: cis and trans in the balance.FEBS Lett. 2009 Dec 17;583(24):3959-65. doi: 10.1016/j.febslet.2009.11.032. FEBS Lett. 2009. PMID: 19914250 Free PMC article. Review.
Cited by
-
Lineage-specific selection and the evolution of virulence in the Candida clade.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2016818118. doi: 10.1073/pnas.2016818118. Proc Natl Acad Sci U S A. 2021. PMID: 33723044 Free PMC article.
-
Environmentally robust cis-regulatory changes underlie rapid climatic adaptation.Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2214614120. doi: 10.1073/pnas.2214614120. Epub 2023 Sep 19. Proc Natl Acad Sci U S A. 2023. PMID: 37725649 Free PMC article.
-
Diversity and Postzygotic Evolution of the Mitochondrial Genome in Hybrids of Saccharomyces Species Isolated by Double Sterility Barrier.Front Microbiol. 2020 May 7;11:838. doi: 10.3389/fmicb.2020.00838. eCollection 2020. Front Microbiol. 2020. PMID: 32457720 Free PMC article.
-
Population and comparative genetics of thermotolerance divergence between yeast species.G3 (Bethesda). 2021 Jul 14;11(7):jkab139. doi: 10.1093/g3journal/jkab139. G3 (Bethesda). 2021. PMID: 33914073 Free PMC article.
-
Divergence in the Saccharomyces Species' Heat Shock Response Is Indicative of Their Thermal Tolerance.Genome Biol Evol. 2023 Nov 1;15(11):evad207. doi: 10.1093/gbe/evad207. Genome Biol Evol. 2023. PMID: 37972247 Free PMC article.
References
-
- Becker B, Feller A, El Alami M, Dubois E, Piérard A.. 1998. A nonameric core sequence is required upstream of the LYSgenes of Saccharomyces cerevisiaefor Lys14p-mediated activation and apparent repression by lysine. Mol Microbiol. 29:151–163. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

