Reconstitution of a functional 7SK snRNP
- PMID: 28431135
- PMCID: PMC5499737
- DOI: 10.1093/nar/gkx262
Reconstitution of a functional 7SK snRNP
Abstract
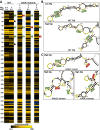
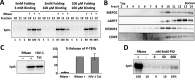
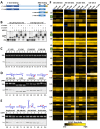
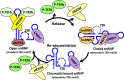
The 7SK small nuclear ribonucleoprotein (snRNP) plays a central role in RNA polymerase II elongation control by regulating the availability of active P-TEFb. We optimized conditions for analyzing 7SK RNA by SHAPE and demonstrated a hysteretic effect of magnesium on 7SK folding dynamics including a 7SK GAUC motif switch. We also found evidence that the 5΄ end pairs alternatively with two different regions of 7SK giving rise to open and closed forms that dictate the state of the 7SK motif. We then used recombinant P-TEFb, HEXIM1, LARP7 and MEPCE to reconstruct a functional 7SK snRNP in vitro. Stably associated P-TEFb was highly inhibited, but could still be released and activated by HIV-1 Tat. Notably, P-TEFb association with both in vitro-reconstituted and cellular snRNPs led to similar changes in SHAPE reactivities, confirming that 7SK undergoes a P-TEFb-dependent structural change. We determined that the xRRM of LARP7 binds to the 3΄ stem loop of 7SK and inhibits the methyltransferase activity of MEPCE through a C-terminal MEPCE interaction domain (MID). Inhibition of MEPCE is dependent on the structure of the 3΄ stem loop and the closed form of 7SK RNA. This study provides important insights into intramolecular interactions within the 7SK snRNP.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP.Nucleic Acids Res. 2013 Apr;41(8):4686-98. doi: 10.1093/nar/gkt159. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23471002 Free PMC article.
-
hLARP7 C-terminal domain contains an xRRM that binds the 3' hairpin of 7SK RNA.Nucleic Acids Res. 2016 Nov 16;44(20):9977-9989. doi: 10.1093/nar/gkw833. Epub 2016 Sep 26. Nucleic Acids Res. 2016. PMID: 27679474 Free PMC article.
-
A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP.Nucleic Acids Res. 2010 Jan;38(2):360-9. doi: 10.1093/nar/gkp977. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906723 Free PMC article.
-
The 7SK snRNP complex: a critical regulator in carcinogenesis.Biochimie. 2025 May 12:S0300-9084(25)00084-7. doi: 10.1016/j.biochi.2025.05.003. Online ahead of print. Biochimie. 2025. PMID: 40368082 Review.
-
Stabilize and connect: the role of LARP7 in nuclear non-coding RNA metabolism.RNA Biol. 2021 Feb;18(2):290-303. doi: 10.1080/15476286.2020.1767952. Epub 2020 Jun 3. RNA Biol. 2021. PMID: 32401147 Free PMC article. Review.
Cited by
-
Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):E6457-E6466. doi: 10.1073/pnas.1806276115. Epub 2018 Jun 26. Proc Natl Acad Sci U S A. 2018. PMID: 29946027 Free PMC article.
-
Visualizing a two-state conformational ensemble in stem-loop 3 of the transcriptional regulator 7SK RNA.Nucleic Acids Res. 2024 Jan 25;52(2):940-952. doi: 10.1093/nar/gkad1159. Nucleic Acids Res. 2024. PMID: 38084902 Free PMC article.
-
An alternative D. melanogaster 7SK snRNP.BMC Mol Cell Biol. 2021 Aug 31;22(1):43. doi: 10.1186/s12860-021-00381-7. BMC Mol Cell Biol. 2021. PMID: 34461828 Free PMC article.
-
Catalytic activity of the Bin3/MePCE methyltransferase domain is dispensable for 7SK snRNP function in Drosophila melanogaster.Genetics. 2024 Jan 3;226(1):iyad203. doi: 10.1093/genetics/iyad203. Genetics. 2024. PMID: 37982586 Free PMC article.
-
Design, synthesis, and biological evaluation of AV6 derivatives as novel dual reactivators of latent HIV-1.RSC Adv. 2018 May 11;8(31):17279-17292. doi: 10.1039/c8ra01216d. eCollection 2018 May 9. RSC Adv. 2018. PMID: 35539279 Free PMC article.
References
-
- Peterlin B.M., Price D.H.. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006; 23:297–305. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

