A reference floral transcriptome of sexual and apomictic Paspalum notatum
- PMID: 28431521
- PMCID: PMC5399859
- DOI: 10.1186/s12864-017-3700-z
A reference floral transcriptome of sexual and apomictic Paspalum notatum
Abstract
Background: Paspalum notatum Flügge is a subtropical grass native to South America, which includes sexual diploid and apomictic polyploid biotypes. In the past decade, a number of apomixis-associated genes were discovered in this species through genetic mapping and differential expression surveys. However, the scarce information on Paspalum sequences available in public databanks limited annotations and functional predictions for these candidates.
Results: We used a long-read 454/Roche FLX+ sequencing strategy to produce robust reference transcriptome datasets from florets of sexual and apomictic Paspalum notatum genotypes and delivered a list of transcripts showing differential representation in both reproductive types. Raw data originated from floral samples collected from premeiosis to anthesis was assembled in three libraries: i) sexual (SEX), ii) apomictic (APO) and iii) global (SEX + APO). A group of physically-supported Paspalum mRNA and EST sequences matched with high level of confidence to both sexual and apomictic libraries. A preliminary trial allowed discovery of the whole set of putative alleles/paralogs corresponding to 23 previously identified apomixis-associated candidate genes. Moreover, a list of 3,732 transcripts and several co-expression and protein -protein interaction networks associated with apomixis were identified.
Conclusions: The use of the 454/Roche FLX+ transcriptome database will allow the detailed characterization of floral alleles/paralogs of apomixis candidate genes identified in prior and future work. Moreover, it was used to reveal additional candidate genes differentially represented in apomictic and sexual flowers. Gene ontology (GO) analyses of this set of transcripts indicated that the main molecular pathways altered in the apomictic genotype correspond to specific biological processes, like biotic and abiotic stress responses, growth, development, cell death and senescence. This data collection will be of interest to the plant reproduction research community and, particularly, to Paspalum breeding projects.
Keywords: Apomixis; Next generation sequencing; Plant reproduction; Sexual reproduction; Transcriptomics.
Figures

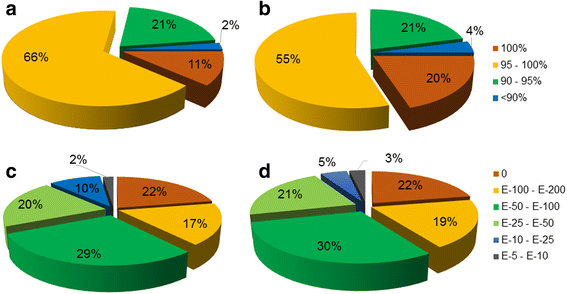

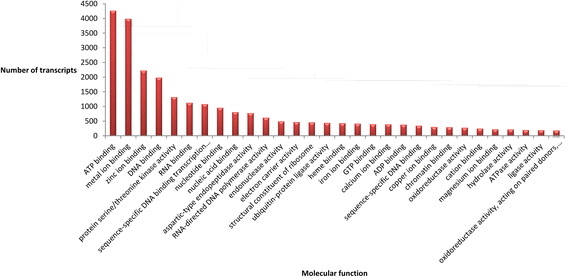
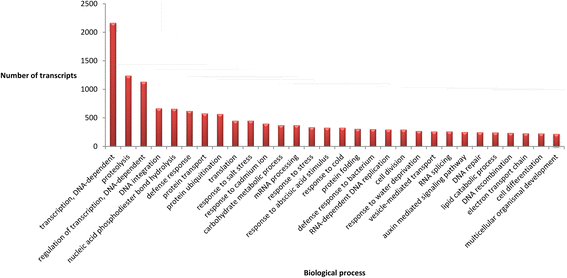


References
-
- Asker SE, Jerling L. Apomixis in plants. Boca Raton: CRC Press; 1982.
-
- Curtis MD, Grossniklaus U. Molecular control of autonomous embryo and endosperm development. Sex Plant Reprod. 2008;21:79–88. doi: 10.1007/s00497-007-0061-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

