Transcriptome profiling of Elymus sibiricus, an important forage grass in Qinghai-Tibet plateau, reveals novel insights into candidate genes that potentially connected to seed shattering
- PMID: 28431567
- PMCID: PMC5399857
- DOI: 10.1186/s12870-017-1026-2
Transcriptome profiling of Elymus sibiricus, an important forage grass in Qinghai-Tibet plateau, reveals novel insights into candidate genes that potentially connected to seed shattering
Abstract
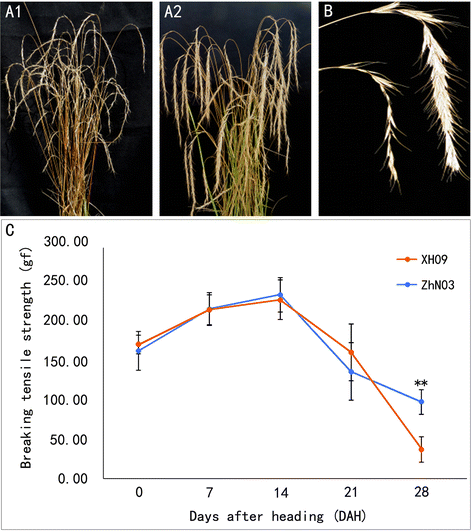
Background: Elymus sibiricus is an important forage grass in semi-arid regions, but it is difficult to grow for commercial seed production due to high seed shattering. To better understand the underlying mechanism and explore the putative genes related to seed shattering, we conducted a combination of morphological, histological, physiochemical and transcriptome analysis on two E. sibiricus genotypes (XH09 and ZhN03) that have contrasting seed shattering.
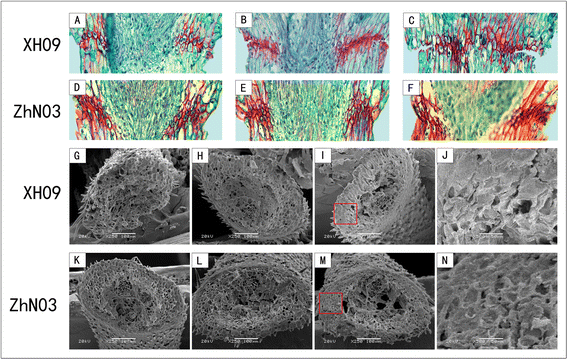
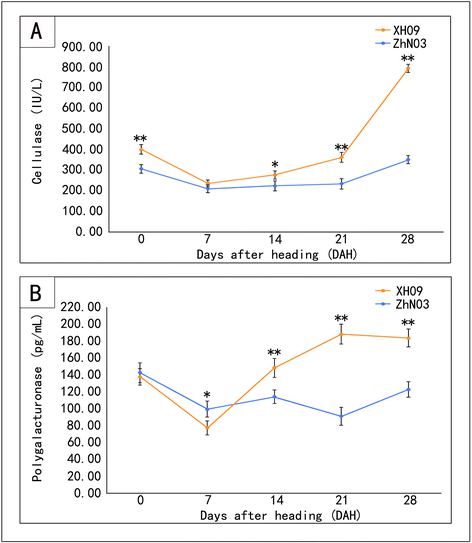
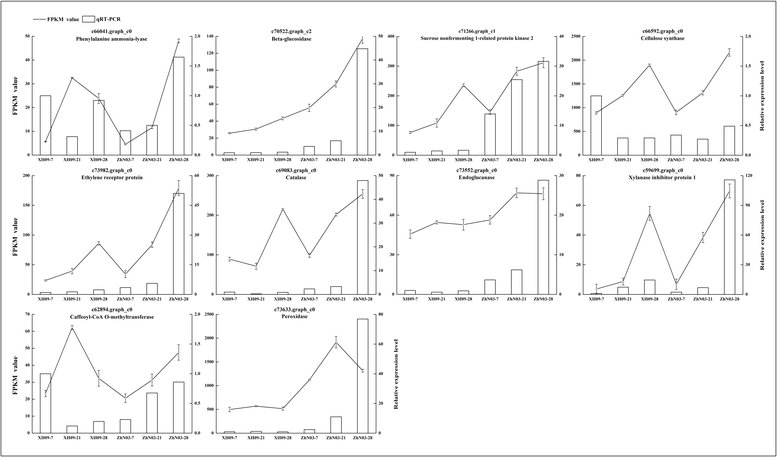
Results: The results show that seed shattering is generally caused by a degradation of the abscission layer. Early degradation of abscission layers was associated with the increased seed shattering in high seed shattering genotype XH09. Two cell wall degrading enzymes, cellulase (CE) and polygalacturonase (PG), had different activity in the abscission zone, indicating their roles in differentiation of abscission layer. cDNA libraries from abscission zone tissue of XH09 and ZhN03 at 7 days, 21 days and 28 days after heading were constructed and sequenced. A total of 86,634 unigenes were annotated and 7110 differentially expressed transcripts (DETs) were predicted from "XH09-7 vs ZhN03-7", "XH09-21 vs ZhN03-21" and "XH09-28 vs ZhN03-28", corresponding to 2058 up-regulated and 5052 down-regulated unigenes. The expression profiles of 10 candidate transcripts involved in cell wall-degrading enzymes, lignin biosynthesis and phytohormone activity were validated using quantitative real-time PCR (qRT-PCR), 8 of which were up-regulated in low seed shattering genotype ZhN03, suggesting these genes may be associated with reduction of seed shattering.
Conclusions: The expression data generated in this study provides an important resource for future molecular biological research in E. sibiricus.
Keywords: Abscission layers; Elymus sibiricus; Mechanism; Next-generation sequencing; Seed shattering; Transcriptome analysis.
Figures

References
-
- Dong Y, Wang YZ. Seed shattering: from models to crops. Front Plant Sci. 2015;6:476. Available from: https://www.ncbi.nlm.nih.gov/pubmed/2615745. Accessed 7 May 2016. - PMC - PubMed
-
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. Available from: https://www.ncbi.nlm.nih.gov/pubmed/16614172. Accessed 7 May 2016. - PubMed
-
- Patterson SE. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. Available from: https://www.ncbi.nlm.nih.gov/pubmed/11402180. Accessed 7 May 2016. - PMC - PubMed
-
- Elgersma A, Leeuwangh JE, Wilms HJ. Abscission and seed shattering in perennial ryegrass (Lolium perenne L.). Euphytica. 1988;39:51–57. Available from: https://link.springer.com/article/10.1007%2FBF00043367. Accessed 7 May 2016. - DOI
-
- Thurber CS, Hepler PK, Caicedo AL. Timing is everything: early degradation of abscission layer is associated with increased seed shattering in U.S. weedy rice. BMC Plant Biol. 2011;11:1–10. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21235796. Accessed 7 May 2016. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

