Centrin diversity and basal body patterning across evolution: new insights from Paramecium
- PMID: 28432105
- PMCID: PMC5483020
- DOI: 10.1242/bio.024273
Centrin diversity and basal body patterning across evolution: new insights from Paramecium
Abstract
First discovered in unicellular eukaryotes, centrins play crucial roles in basal body duplication and anchoring mechanisms. While the evolutionary status of the founding members of the family, Centrin2/Vfl2 and Centrin3/cdc31 has long been investigated, the evolutionary origin of other members of the family has received less attention. Using a phylogeny of ciliate centrins, we identify two other centrin families, the ciliary centrins and the centrins present in the contractile filaments (ICL centrins). In this paper, we carry on the functional analysis of still not well-known centrins, the ICL1e subfamily identified in Paramecium, and show their requirement for correct basal body anchoring through interactions with Centrin2 and Centrin3. Using Paramecium as well as a eukaryote-wide sampling of centrins from completely sequenced genomes, we revisited the evolutionary story of centrins. Their phylogeny shows that the centrins associated with the ciliate contractile filaments are widespread in eukaryotic lineages and could be as ancient as Centrin2 and Centrin3.
Keywords: Basal body anchoring; Basal body assembly; Centrin evolution; Ciliary centrins; Ciliated epithelia polarity.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


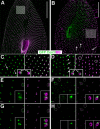
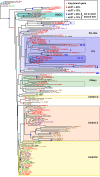
Similar articles
-
Functional diversification of centrins and cell morphological complexity.J Cell Sci. 2008 Jan 1;121(Pt 1):65-74. doi: 10.1242/jcs.019414. Epub 2007 Dec 4. J Cell Sci. 2008. PMID: 18057024
-
Centrin deficiency in Paramecium affects the geometry of basal-body duplication.Curr Biol. 2005 Dec 6;15(23):2097-106. doi: 10.1016/j.cub.2005.11.038. Curr Biol. 2005. PMID: 16332534
-
A Centrin3-dependent, transient, appendage of the mother basal body guides the positioning of the daughter basal body in Paramecium.Protist. 2013 May;164(3):352-68. doi: 10.1016/j.protis.2012.11.003. Epub 2012 Dec 20. Protist. 2013. PMID: 23261281
-
Such small hands: the roles of centrins/caltractins in the centriole and in genome maintenance.Cell Mol Life Sci. 2012 Sep;69(18):2979-97. doi: 10.1007/s00018-012-0961-1. Epub 2012 Mar 30. Cell Mol Life Sci. 2012. PMID: 22460578 Free PMC article. Review.
-
Structural Basis for the Functional Diversity of Centrins: A Focus on Calcium Sensing Properties and Target Recognition.Int J Mol Sci. 2021 Nov 10;22(22):12173. doi: 10.3390/ijms222212173. Int J Mol Sci. 2021. PMID: 34830049 Free PMC article. Review.
Cited by
-
Using Paramecium as a Model for Ciliopathies.Genes (Basel). 2021 Sep 24;12(10):1493. doi: 10.3390/genes12101493. Genes (Basel). 2021. PMID: 34680887 Free PMC article. Review.
-
The Cilioprotist Cytoskeleton , a Model for Understanding How Cell Architecture and Pattern Are Specified: Recent Discoveries from Ciliates and Comparable Model Systems.Methods Mol Biol. 2022;2364:251-295. doi: 10.1007/978-1-0716-1661-1_13. Methods Mol Biol. 2022. PMID: 34542858 Review.
-
Decoding ultrasensitive self-assembly of the calcium-regulated Tetrahymena cytoskeletal protein Tcb2 using optical actuation.bioRxiv [Preprint]. 2025 May 29:2025.05.26.656216. doi: 10.1101/2025.05.26.656216. bioRxiv. 2025. PMID: 40501648 Free PMC article. Preprint.
-
Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota.Nat Commun. 2021 Aug 17;12(1):4973. doi: 10.1038/s41467-021-25308-w. Nat Commun. 2021. PMID: 34404788 Free PMC article.
-
A polarized multicomponent foundation upholds ciliary central microtubules.J Mol Cell Biol. 2025 Jan 30;16(8):mjae031. doi: 10.1093/jmcb/mjae031. J Mol Cell Biol. 2025. PMID: 39165107 Free PMC article.
References
-
- Aubusson-Fleury A., Lemullois M., Garreau de Loubresse N., Laligné C., Cohen J., Rosnet O., Jerka-Dziadosz M., Beisson J. and Koll F. (2012). The conserved centrosomal protein FOR20 is required for assembly of the transition zone and basal body docking at the cell surface. J. Cell Sci. 125, 4395-4404. 10.1242/jcs.108639 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

