N-terminal Proteomics Assisted Profiling of the Unexplored Translation Initiation Landscape in Arabidopsis thaliana
- PMID: 28432195
- PMCID: PMC5461538
- DOI: 10.1074/mcp.M116.066662
N-terminal Proteomics Assisted Profiling of the Unexplored Translation Initiation Landscape in Arabidopsis thaliana
Abstract
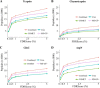
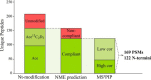
Proteogenomics is an emerging research field yet lacking a uniform method of analysis. Proteogenomic studies in which N-terminal proteomics and ribosome profiling are combined, suggest that a high number of protein start sites are currently missing in genome annotations. We constructed a proteogenomic pipeline specific for the analysis of N-terminal proteomics data, with the aim of discovering novel translational start sites outside annotated protein coding regions. In summary, unidentified MS/MS spectra were matched to a specific N-terminal peptide library encompassing protein N termini encoded in the Arabidopsis thaliana genome. After a stringent false discovery rate filtering, 117 protein N termini compliant with N-terminal methionine excision specificity and indicative of translation initiation were found. These include N-terminal protein extensions and translation from transposable elements and pseudogenes. Gene prediction provided supporting protein-coding models for approximately half of the protein N termini. Besides the prediction of functional domains (partially) contained within the newly predicted ORFs, further supporting evidence of translation was found in the recently released Araport11 genome re-annotation of Arabidopsis and computational translations of sequences stored in public repositories. Most interestingly, complementary evidence by ribosome profiling was found for 23 protein N termini. Finally, by analyzing protein N-terminal peptides, an in silico analysis demonstrates the applicability of our N-terminal proteogenomics strategy in revealing protein-coding potential in species with well- and poorly-annotated genomes.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Ribosome signatures aid bacterial translation initiation site identification.BMC Biol. 2017 Aug 30;15(1):76. doi: 10.1186/s12915-017-0416-0. BMC Biol. 2017. PMID: 28854918 Free PMC article.
-
Discovery and revision of Arabidopsis genes by proteogenomics.Proc Natl Acad Sci U S A. 2008 Dec 30;105(52):21034-8. doi: 10.1073/pnas.0811066106. Epub 2008 Dec 19. Proc Natl Acad Sci U S A. 2008. PMID: 19098097 Free PMC article.
-
Improved super-resolution ribosome profiling reveals prevalent translation of upstream ORFs and small ORFs in Arabidopsis.Plant Cell. 2024 Feb 26;36(3):510-539. doi: 10.1093/plcell/koad290. Plant Cell. 2024. PMID: 38000896 Free PMC article.
-
False discovery rate: the Achilles' heel of proteogenomics.Brief Bioinform. 2022 Sep 20;23(5):bbac163. doi: 10.1093/bib/bbac163. Brief Bioinform. 2022. PMID: 35534181 Review.
-
Bacterial riboproteogenomics: the era of N-terminal proteoform existence revealed.FEMS Microbiol Rev. 2020 Jul 1;44(4):418-431. doi: 10.1093/femsre/fuaa013. FEMS Microbiol Rev. 2020. PMID: 32386204 Review.
Cited by
-
The Arabidopsis PeptideAtlas: Harnessing worldwide proteomics data to create a comprehensive community proteomics resource.Plant Cell. 2021 Nov 4;33(11):3421-3453. doi: 10.1093/plcell/koab211. Plant Cell. 2021. PMID: 34411258 Free PMC article.
-
Plant genome information facilitates plant functional genomics.Planta. 2024 Apr 9;259(5):117. doi: 10.1007/s00425-024-04397-z. Planta. 2024. PMID: 38592421 Free PMC article. Review.
-
Charting the N-Terminal Acetylome: A Comprehensive Map of Human NatA Substrates.Int J Mol Sci. 2021 Oct 2;22(19):10692. doi: 10.3390/ijms221910692. Int J Mol Sci. 2021. PMID: 34639033 Free PMC article.
-
A novel short L-arginine responsive protein-coding gene (laoB) antiparallel overlapping to a CadC-like transcriptional regulator in Escherichia coli O157:H7 Sakai originated by overprinting.BMC Evol Biol. 2018 Feb 12;18(1):21. doi: 10.1186/s12862-018-1134-0. BMC Evol Biol. 2018. PMID: 29433444 Free PMC article.
-
Lost and Found: Re-searching and Re-scoring Proteomics Data Aids Genome Annotation and Improves Proteome Coverage.mSystems. 2020 Oct 27;5(5):e00833-20. doi: 10.1128/mSystems.00833-20. mSystems. 2020. PMID: 33109751 Free PMC article.
References
-
- Jaffe J. D., Berg H. C., and Church G. M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 - PubMed
-
- Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., Liebler D. C., and NCI CPTAC (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 - PMC - PubMed
-
- Ruggles K. V., Tang Z., Wang X., Grover H., Askenazi M., Teubl J., Cao S., McLellan M. D., Clauser K. R., Tabb D. L., Mertins P., Slebos R., Erdmann-Gilmore P., Li S., Gunawardena H. P., Xie L., Liu T., Zhou J. Y., Sun S., Hoadley K. A., Perou C. M., Chen X., Davies S. R., Maher C. A., Kinsinger C. R., Rodland K. D., Zhang H., Zhang Z., Ding L., Townsend R. R., Rodriguez H., Chan D., Smith R. D., Liebler D. C., Carr S. A., Payne S., Ellis M. J., and Fenyo D. (2016) An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol. Cell. Proteomics 15, 1060–1071 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

