Mechanistic insights into epigenetic modulation of ethanol consumption
- PMID: 28433417
- PMCID: PMC5550096
- DOI: 10.1016/j.alcohol.2017.01.016
Mechanistic insights into epigenetic modulation of ethanol consumption
Abstract
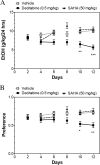
There is growing evidence that small-molecule inhibitors of epigenetic modulators, such as histone deacetylases (HDAC) and DNA methyltransferases (DNMT), can reduce voluntary ethanol consumption in animal models, but molecular and cellular processes underlying this behavioral effect are poorly understood. We used C57BL/6J male mice to investigate the effects of two FDA-approved drugs, decitabine (a DNMT inhibitor) and SAHA (an HDAC inhibitor), on ethanol consumption using two tests: binge-like drinking in the dark (DID) and chronic intermittent every other day (EOD) drinking. Decitabine but not SAHA reduced ethanol consumption in both tests. We further investigated decitabine's effects on the brain's reward pathway by gene expression profiling in the ventral tegmental area (VTA), using RNA sequencing and electrophysiological recordings from VTA dopaminergic neurons. Decitabine-induced decreases in EOD drinking were associated with global changes in gene expression, implicating regulation of cerebral blood flow, extracellular matrix organization, and neuroimmune functions in decitabine actions. In addition, an in vivo administration of decitabine shortened ethanol-induced excitation of VTA dopaminergic neurons in vitro, suggesting that decitabine reduces ethanol drinking via changes in the reward pathway. Taken together, our data suggest a contribution of both neuronal and non-neuronal mechanisms in the VTA in the regulation of ethanol consumption. Decitabine and other epigenetic compounds have been approved for cancer treatment, and understanding their mechanisms of actions in the brain may assist in repurposing these drugs and developing novel therapies for central disorders, including drug addiction.
Keywords: Alcohol; Behavior; Drug repurposing; Epigenetics; Neuronal activity; Transcriptome.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Epigenetic mechanisms are involved in the regulation of ethanol consumption in mice.Int J Neuropsychopharmacol. 2014 Oct 31;18(2):pyu072. doi: 10.1093/ijnp/pyu072. Int J Neuropsychopharmacol. 2014. PMID: 25522411 Free PMC article.
-
[Epigenetic mechanisms and alcohol use disorders: a potential therapeutic target].Biol Aujourdhui. 2017;211(1):83-91. doi: 10.1051/jbio/2017014. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682229 Review. French.
-
Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: reversal by histone deacetylase inhibitors.Neuropsychopharmacology. 2013 Aug;38(9):1674-84. doi: 10.1038/npp.2013.65. Epub 2013 Mar 8. Neuropsychopharmacology. 2013. PMID: 23474591 Free PMC article.
-
Alcohol consumption induces global gene expression changes in VTA dopaminergic neurons.Genes Brain Behav. 2016 Mar;15(3):318-26. doi: 10.1111/gbb.12266. Epub 2015 Dec 28. Genes Brain Behav. 2016. PMID: 26482798 Free PMC article.
-
Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine.Int J Cancer. 2008 Jul 1;123(1):8-13. doi: 10.1002/ijc.23607. Int J Cancer. 2008. PMID: 18425818 Review.
Cited by
-
Evidence for Modulation of Substance Use Disorders by the Gut Microbiome: Hidden in Plain Sight.Pharmacol Rev. 2021 Apr;73(2):571-596. doi: 10.1124/pharmrev.120.000144. Pharmacol Rev. 2021. PMID: 33597276 Free PMC article. Review.
-
Modulation of neuronal excitability by binge alcohol drinking.Front Mol Neurosci. 2023 Feb 14;16:1098211. doi: 10.3389/fnmol.2023.1098211. eCollection 2023. Front Mol Neurosci. 2023. PMID: 36866357 Free PMC article. Review.
-
DNA modifications in models of alcohol use disorders.Alcohol. 2017 May;60:19-30. doi: 10.1016/j.alcohol.2016.11.004. Epub 2016 Nov 9. Alcohol. 2017. PMID: 27865607 Free PMC article. Review.
-
Epigenetic drugs and psychedelics as emerging therapies for alcohol use disorder: insights from preclinical studies.J Neural Transm (Vienna). 2024 May;131(5):525-561. doi: 10.1007/s00702-024-02757-3. Epub 2024 Mar 30. J Neural Transm (Vienna). 2024. PMID: 38554193 Review.
-
Ethanol actions on the ventral tegmental area: novel potential targets on reward pathway neurons.Psychopharmacology (Berl). 2018 Jun;235(6):1711-1726. doi: 10.1007/s00213-018-4875-y. Epub 2018 Mar 16. Psychopharmacology (Berl). 2018. PMID: 29549390 Free PMC article. Review.
References
-
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Current Opinion in Pharmacology. 2008;8(1):57–64. http://doi.org/10.1016/j.coph.2007.12.002. - DOI - PMC - PubMed
-
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, et al. DNA Methylation in the Medial Prefrontal Cortex Regulates Alcohol-Induced Behavior and Plasticity. Journal of Neuroscience. 2015;35(15):6153–6164. http://doi.org/10.1523/JNEUROSCI.4571-14.2015. - DOI - PMC - PubMed
-
- Berridge KC, Robinson TE. What is the role of dopamine in reward: hedonic impact, reward learning, or incentive salience? Brain Research Brain Research Reviews. 1998;28(3):309–369. - PubMed
-
- Brodie MS, Appel SB. The effects of ethanol on dopaminergic neurons of the ventral tegmental area studied with intracellular recording in brain slices. Alcoholism: Clinical and Experimental Research. 1998;22(1):236–244. - PubMed
-
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. http://doi.org/10.1186/1471-2105-14-128. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

