Prefrontal cortex expression of chromatin modifier genes in male WSP and WSR mice changes across ethanol dependence, withdrawal, and abstinence
- PMID: 28433423
- PMCID: PMC5497775
- DOI: 10.1016/j.alcohol.2017.01.010
Prefrontal cortex expression of chromatin modifier genes in male WSP and WSR mice changes across ethanol dependence, withdrawal, and abstinence
Abstract
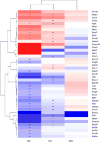
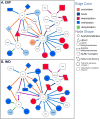
Alcohol-use disorder (AUD) is a relapsing disorder associated with excessive ethanol consumption. Recent studies support the involvement of epigenetic mechanisms in the development of AUD. Studies carried out so far have focused on a few specific epigenetic modifications. The goal of this project was to investigate gene expression changes of epigenetic regulators that mediate a broad array of chromatin modifications after chronic alcohol exposure, chronic alcohol exposure followed by 8 h withdrawal, and chronic alcohol exposure followed by 21 days of abstinence in Withdrawal-Resistant (WSR) and Withdrawal Seizure-Prone (WSP) selected mouse lines. We found that chronic vapor exposure to highly intoxicating levels of ethanol alters the expression of several chromatin remodeling genes measured by quantitative PCR array analyses. The identified effects were independent of selected lines, which, however, displayed baseline differences in epigenetic gene expression. We reported dysregulation in the expression of genes involved in histone acetylation, deacetylation, lysine and arginine methylation and ubiquitinationhylation during chronic ethanol exposure and withdrawal, but not after 21 days of abstinence. Ethanol-induced changes are consistent with decreased histone acetylation and with decreased deposition of the permissive ubiquitination mark H2BK120ub, associated with reduced transcription. On the other hand, ethanol-induced changes in the expression of genes involved in histone lysine methylation are consistent with increased transcription. The net result of these modifications on gene expression is likely to depend on the combination of the specific histone tail modifications present at a given time on a given promoter. Since alcohol does not modulate gene expression unidirectionally, it is not surprising that alcohol does not unidirectionally alter chromatin structure toward a closed or open state, as suggested by the results of this study.
Keywords: Alcohol-use disorder; DNA methylation; Epigenetics; Histone acetylation; Histone arginine acetylation; Histone lysine methylation; Histone ubiquitination.
Published by Elsevier Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

Similar articles
-
The Impact of Alcohol-Induced Epigenetic Modifications in the Treatment of Alcohol use Disorders.Curr Med Chem. 2024;31(36):5837-5855. doi: 10.2174/0109298673256937231004093143. Curr Med Chem. 2024. PMID: 37828672 Review.
-
[Epigenetic mechanisms and alcohol use disorders: a potential therapeutic target].Biol Aujourdhui. 2017;211(1):83-91. doi: 10.1051/jbio/2017014. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682229 Review. French.
-
Epigenetic mechanisms are involved in the regulation of ethanol consumption in mice.Int J Neuropsychopharmacol. 2014 Oct 31;18(2):pyu072. doi: 10.1093/ijnp/pyu072. Int J Neuropsychopharmacol. 2014. PMID: 25522411 Free PMC article.
-
Importance of genetic background for risk of relapse shown in altered prefrontal cortex gene expression during abstinence following chronic alcohol intoxication.Neuroscience. 2011 Jan 26;173:57-75. doi: 10.1016/j.neuroscience.2010.11.006. Epub 2010 Nov 25. Neuroscience. 2011. PMID: 21081154 Free PMC article.
-
Correlation between the epigenetic modification of histone H3K9 acetylation of NR2B gene promoter in rat hippocampus and ethanol withdrawal syndrome.Mol Biol Rep. 2019 Jun;46(3):2867-2875. doi: 10.1007/s11033-019-04733-7. Epub 2019 Mar 22. Mol Biol Rep. 2019. PMID: 30903572
Cited by
-
Histone Methylation Regulation in Neurodegenerative Disorders.Int J Mol Sci. 2021 Apr 28;22(9):4654. doi: 10.3390/ijms22094654. Int J Mol Sci. 2021. PMID: 33925016 Free PMC article. Review.
-
Effects of repeated alcohol abstinence on within-subject prefrontal cortical gene expression in rhesus macaques.Adv Drug Alcohol Res. 2024 Apr 26;4:12528. doi: 10.3389/adar.2024.12528. eCollection 2024. Adv Drug Alcohol Res. 2024. PMID: 38737578 Free PMC article.
-
Histone Deacetylase Inhibitor Suberanilohydroxamic Acid Treatment Reverses Hyposensitivity to γ-Aminobutyric Acid in the Ventral Tegmental Area During Ethanol Withdrawal.Alcohol Clin Exp Res. 2018 Nov;42(11):2160-2171. doi: 10.1111/acer.13870. Epub 2018 Sep 7. Alcohol Clin Exp Res. 2018. PMID: 30103280 Free PMC article.
-
Epigenetic mediators and consequences of excessive alcohol consumption.Alcohol. 2017 May;60:1-6. doi: 10.1016/j.alcohol.2017.02.357. Epub 2017 Mar 11. Alcohol. 2017. PMID: 28395929 Free PMC article. Review. No abstract available.
-
Neonatal Alcohol Exposure in Mice Induces Select Differentiation- and Apoptosis-Related Chromatin Changes Both Independent of and Dependent on Sex.Front Genet. 2020 Feb 11;11:35. doi: 10.3389/fgene.2020.00035. eCollection 2020. Front Genet. 2020. PMID: 32117449 Free PMC article.
References
-
- Beadles-Bohling A, Wiren K. Anticonvulsive effects of kappa opioid receptor modulation in an animal model of ethanol withdrawal. Genes Brain Behav. 2006;5:483–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

