Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma
- PMID: 28434841
- PMCID: PMC5462626
- DOI: 10.1016/j.ccell.2017.03.011
Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma
Abstract
Diffuse intrinsic pontine glioma (DIPG) is a fatal pediatric cancer with limited therapeutic options. The majority of cases of DIPG exhibit a mutation in histone-3 (H3K27M) that results in oncogenic transcriptional aberrancies. We show here that DIPG is vulnerable to transcriptional disruption using bromodomain inhibition or CDK7 blockade. Targeting oncogenic transcription through either of these methods synergizes with HDAC inhibition, and DIPG cells resistant to HDAC inhibitor therapy retain sensitivity to CDK7 blockade. Identification of super-enhancers in DIPG provides insights toward the cell of origin, highlighting oligodendroglial lineage genes, and reveals unexpected mechanisms mediating tumor viability and invasion, including potassium channel function and EPH receptor signaling. The findings presented demonstrate transcriptional vulnerabilities and elucidate previously unknown mechanisms of DIPG pathobiology.
Keywords: BRD4; CDK7; DIPG; EPH; oligodendrocyte precursor cell; potassium channel; super-enhancer.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
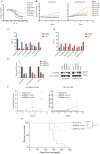
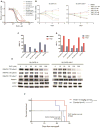
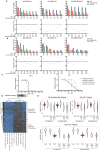

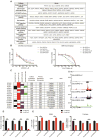
Comment in
-
Using Epigenetic Reprogramming to Treat Pediatric Brain Cancer.Cancer Cell. 2017 May 8;31(5):609-611. doi: 10.1016/j.ccell.2017.04.008. Cancer Cell. 2017. PMID: 28486100
References
-
- Araki T, et al. Mechanism of Homophilic Binding Mediated by Ninjurin, a Novel Widely Expressed Adhesion Molecule. Journal of Biological Chemistry. 1997;272(34):21373–21380. - PubMed
-
- Araki T, Milbrandt J. Ninjurin, a Novel Adhesion Molecule, Is Induced by Nerve Injury and Promotes Axonal Growth. Neuron. 1996;17(2):353–361. - PubMed
-
- Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer cell. 2013;24(5):660–72. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

