Prognostic Biomarker Identification Through Integrating the Gene Signatures of Hepatocellular Carcinoma Properties
- PMID: 28434945
- PMCID: PMC5440601
- DOI: 10.1016/j.ebiom.2017.04.014
Prognostic Biomarker Identification Through Integrating the Gene Signatures of Hepatocellular Carcinoma Properties
Abstract
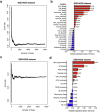
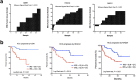
Many molecular classification and prognostic gene signatures for hepatocellular carcinoma (HCC) patients have been established based on genome-wide gene expression profiling; however, their generalizability is unclear. Herein, we systematically assessed the prognostic effects of these gene signatures and identified valuable prognostic biomarkers by integrating these gene signatures. With two independent HCC datasets (GSE14520, N=242 and GSE54236, N=78), 30 published gene signatures were evaluated, and 11 were significantly associated with the overall survival (OS) of postoperative HCC patients in both datasets. The random survival forest models suggested that the gene signatures were superior to clinical characteristics for predicting the prognosis of the patients. Based on the 11 gene signatures, a functional protein-protein interaction (PPI) network with 1406 nodes and 10,135 edges was established. With tissue microarrays of HCC patients (N=60), we determined the prognostic values of the core genes in the network and found that RAD21, CDK1, and HDAC2 expression levels were negatively associated with OS for HCC patients. The multivariate Cox regression analyses suggested that CDK1 was an independent prognostic factor, which was validated in an independent case cohort (N=78). In cellular models, inhibition of CDK1 by siRNA or a specific inhibitor, RO-3306, reduced cellular proliferation and viability for HCC cells. These results suggest that the prognostic predictive capacities of these gene signatures are reproducible and that CDK1 is a potential prognostic biomarker or therapeutic target for HCC patients.
Keywords: Biomarker; Gene signature; Hepatocellular carcinoma; Network; Overall survival.
Copyright © 2017 The Author(s). Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Assenov Y., Ramirez F., Schelhorn S.E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. - PubMed
-
- Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. - PubMed
-
- Boyault S., Rickman D.S., de Reynies A., Balabaud C., Rebouissou S., Jeannot E., Herault A., Saric J., Belghiti J., Franco D. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. - PubMed
-
- Budhu A., Forgues M., Ye Q.H., Jia H.L., He P., Zanetti K.A., Kammula U.S., Chen Y., Qin L.X., Tang Z.Y. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

