Genetic interrelations in the actinomycin biosynthetic gene clusters of Streptomyces antibioticus IMRU 3720 and Streptomyces chrysomallus ATCC11523, producers of actinomycin X and actinomycin C
- PMID: 28435299
- PMCID: PMC5391158
- DOI: 10.2147/AABC.S117707
Genetic interrelations in the actinomycin biosynthetic gene clusters of Streptomyces antibioticus IMRU 3720 and Streptomyces chrysomallus ATCC11523, producers of actinomycin X and actinomycin C
Abstract
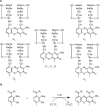
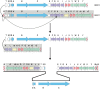
Sequencing the actinomycin (acm) biosynthetic gene cluster of Streptomyces antibioticus IMRU 3720, which produces actinomycin X (Acm X), revealed 20 genes organized into a highly similar framework as in the bi-armed acm C biosynthetic gene cluster of Streptomyces chrysomallus but without an attached additional extra arm of orthologues as in the latter. Curiously, the extra arm of the S. chrysomallus gene cluster turned out to perfectly match the single arm of the S. antibioticus gene cluster in the same order of orthologues including the the presence of two pseudogenes, scacmM and scacmN, encoding a cytochrome P450 and its ferredoxin, respectively. Orthologues of the latter genes were both missing in the principal arm of the S. chrysomallus acm C gene cluster. All orthologues of the extra arm showed a G +C-contents different from that of their counterparts in the principal arm. Moreover, the similarities of translation products from the extra arm were all higher to the corresponding translation products of orthologue genes from the S. antibioticus acm X gene cluster than to those encoded by the principal arm of their own gene cluster. This suggests that the duplicated structure of the S. chrysomallus acm C biosynthetic gene cluster evolved from previous fusion between two one-armed acm gene clusters each from a different genetic background. However, while scacmM and scacmN in the extra arm of the S. chrysomallus acm C gene cluster are mutated and therefore are non-functional, their orthologues saacmM and saacmN in the S. antibioticus acm C gene cluster show no defects seemingly encoding active enzymes with functions specific for Acm X biosynthesis. Both acm biosynthetic gene clusters lack a kynurenine-3-monooxygenase gene necessary for biosynthesis of 3-hydroxy-4-methylanthranilic acid, the building block of the Acm chromophore, which suggests participation of a genome-encoded relevant monooxygenase during Acm biosynthesis in both S. chrysomallus and S. antibioticus.
Keywords: 3-hydroxy-4-methylanthranilic acid (4-MHA); Streptomyces anulatus Streptomyces antibioticus; Streptomyces chrysomallus; actinomycin; actinomycin C; actinomycin X; actinomycin halves; biosynthesis; evolution of biosynthetic gene cluster; genetic transmission of biosynthetic gene cluster; genomes.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures




Similar articles
-
Comparison of actinomycin peptide synthetase formation in Streptomyces chrysomallus and Streptomyces antibioticus.J Basic Microbiol. 2019 Feb;59(2):148-157. doi: 10.1002/jobm.201800244. Epub 2018 Sep 20. J Basic Microbiol. 2019. PMID: 30238507
-
Ketonization of Proline Residues in the Peptide Chains of Actinomycins by a 4-Oxoproline Synthase.Chembiochem. 2018 Apr 4;19(7):706-715. doi: 10.1002/cbic.201700666. Epub 2018 Feb 15. Chembiochem. 2018. PMID: 29327817
-
The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry.J Bacteriol. 2010 May;192(10):2583-95. doi: 10.1128/JB.01526-09. Epub 2010 Mar 19. J Bacteriol. 2010. PMID: 20304989 Free PMC article.
-
Genetics of actinomycin C production in Streptomyces chrysomallus.J Bacteriol. 1988 Mar;170(3):1360-8. doi: 10.1128/jb.170.3.1360-1368.1988. J Bacteriol. 1988. PMID: 2449423 Free PMC article.
-
The biosynthetic gene clusters of aminocoumarin antibiotics.Planta Med. 2006 Oct;72(12):1093-9. doi: 10.1055/s-2006-946699. Epub 2006 Jul 25. Planta Med. 2006. PMID: 16868863 Review.
Cited by
-
Harnessing Actinobacteria secondary metabolites for tuberculosis drug discovery: Historical trends, current status and future outlooks.Nat Prod Bioprospect. 2025 Aug 11;15(1):52. doi: 10.1007/s13659-025-00533-8. Nat Prod Bioprospect. 2025. PMID: 40788464 Free PMC article. Review.
-
Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073.Mar Drugs. 2019 Apr 23;17(4):240. doi: 10.3390/md17040240. Mar Drugs. 2019. PMID: 31018504 Free PMC article.
-
Discovery of actinomycin L, a new member of the actinomycin family of antibiotics.Sci Rep. 2022 Feb 18;12(1):2813. doi: 10.1038/s41598-022-06736-0. Sci Rep. 2022. PMID: 35181725 Free PMC article.
-
Genome mining of actinomycin shunt products from Kitasatospora sp. YINM00002.RSC Adv. 2023 Dec 12;13(51):36200-36208. doi: 10.1039/d3ra07277k. eCollection 2023 Dec 8. RSC Adv. 2023. PMID: 38090065 Free PMC article.
-
Current state and future perspectives of cytochrome P450 enzymes for C-H and C=C oxygenation.Synth Syst Biotechnol. 2022 May 8;7(3):887-899. doi: 10.1016/j.synbio.2022.04.009. eCollection 2022 Sep. Synth Syst Biotechnol. 2022. PMID: 35601824 Free PMC article.
References
-
- Keller U, Krengel U, Haese A. Genetic analysis in Streptomyces chrysomallus. J Gen Microbiol. 1985;131(5):1181–1191. - PubMed
-
- Hopwood DA, Bibb MJ, Chater KF, et al. Genetic Manipulation of Streptomyces. A Laboratory Manual. Norwich, UK: John Innes foundation; 1985.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

