Evolutionary dynamics of CRISPR gene drives
- PMID: 28435878
- PMCID: PMC5381957
- DOI: 10.1126/sciadv.1601964
Evolutionary dynamics of CRISPR gene drives
Abstract
The alteration of wild populations has been discussed as a solution to a number of humanity's most pressing ecological and public health concerns. Enabled by the recent revolution in genome editing, clustered regularly interspaced short palindromic repeats (CRISPR) gene drives-selfish genetic elements that can spread through populations even if they confer no advantage to their host organism-are rapidly emerging as the most promising approach. However, before real-world applications are considered, it is imperative to develop a clear understanding of the outcomes of drive release in nature. Toward this aim, we mathematically study the evolutionary dynamics of CRISPR gene drives. We demonstrate that the emergence of drive-resistant alleles presents a major challenge to previously reported constructs, and we show that an alternative design that selects against resistant alleles could greatly improve evolutionary stability. We discuss all results in the context of CRISPR technology and provide insights that inform the engineering of practical gene drive systems.
Keywords: CRISPR; Resistance; evolutionary dynamics; gene drive.
Figures
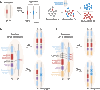
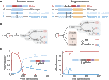
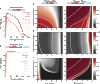
References
-
- Akbari O. S., Bellen H. J., Bier E., Bullock S. L., Burt A., Church G. M., Cook K. R., Duchek P., Edwards O. R., Esvelt K. M., Gantz V. M., Golic K. G., Gratz S. J., Harrison M. M., Hayes K. R., James A. A., Kaufman T. C., Knoblich J., Malik H. S., Matthews K. A., O’Connor-Giles K. M., Parks A. L., Perrimon N., Port F., Russell S., Ueda R., Wildonger J., Safeguarding gene drive experiments in the laboratory. Science 349, 927–929 (2015). - PMC - PubMed
-
- Oye K. A., Esvelt K., Appleton E., Catteruccia F., Church G., Kuiken T., Lightfoot S. B.-Y., McNamara J., Smidler A., Collins J. P., Regulating gene drives. Science 345, 626–628 (2014). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

