Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila
- PMID: 28436983
- PMCID: PMC5484582
- DOI: 10.1038/ng.3848
Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila
Abstract
Transgenerational epigenetic inheritance (TEI) describes the transmission of alternative functional states through multiple generations in the presence of the same genomic DNA sequence. Very little is known about the principles and the molecular mechanisms governing this type of inheritance. Here, by transiently enhancing 3D chromatin interactions, we established stable and isogenic Drosophila epilines that carry alternative epialleles, as defined by differential levels of Polycomb-dependent trimethylation of histone H3 Lys27 (forming H3K27me3). After being established, epialleles can be dominantly transmitted to naive flies and can induce paramutation. Importantly, epilines can be reset to a naive state by disruption of chromatin interactions. Finally, we found that environmental changes modulate the expressivity of the epialleles, and we extended our paradigm to naturally occurring phenotypes. Our work sheds light on how nuclear organization and Polycomb group (PcG) proteins contribute to epigenetically inheritable phenotypic variability.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


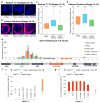



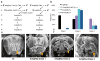
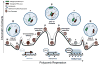
References
-
- Jablonka E, Raz G. Transgenerational Epigenetic Inheritance: prevalence, mechanisms and implications for the study of heredity and evolution. The Quarterly Review Of Biology. 2009;84 - PubMed
-
- Danchin E, et al. Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat Rev Genet. 2011;12:475–86. - PubMed
-
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–62. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

