Role of cleavage at the core-E1 junction of hepatitis C virus polyprotein in viral morphogenesis
- PMID: 28437468
- PMCID: PMC5402940
- DOI: 10.1371/journal.pone.0175810
Role of cleavage at the core-E1 junction of hepatitis C virus polyprotein in viral morphogenesis
Abstract
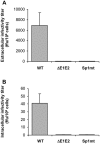
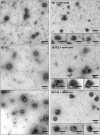
In hepatitis C virus (HCV) polyprotein sequence, core protein terminates with E1 envelope signal peptide. Cleavage by signal peptidase (SP) separates E1 from the complete form of core protein, anchored in the endoplasmic reticulum (ER) membrane by the signal peptide. Subsequent cleavage of the signal peptide by signal-peptide peptidase (SPP) releases the mature form of core protein, which preferentially relocates to lipid droplets. Both of these cleavages are required for the HCV infectious cycle, supporting the idea that HCV assembly begins at the surface of lipid droplets, yet SPP-catalyzed cleavage is dispensable for initiation of budding in the ER. Here we have addressed at what step(s) of the HCV infectious cycle SP-catalyzed cleavage at the core-E1 junction is required. Taking advantage of the sole system that has allowed visualization of HCV budding events in the ER lumen of mammalian cells, we showed that, unexpectedly, mutations abolishing this cleavage did not prevent but instead tended to promote the initiation of viral budding. Moreover, even though no viral particles were released from Huh-7 cells transfected with a full-length HCV genome bearing these mutations, intracellular viral particles containing core protein protected by a membrane envelope were formed. These were visualized by electron microscopy as capsid-containing particles with a diameter of about 70 nm and 40 nm before and after delipidation, respectively, comparable to intracellular wild-type particle precursors except that they were non-infectious. Thus, our results show that SP-catalyzed cleavage is dispensable for HCV budding per se, but is required for the viral particles to acquire their infectivity and secretion. These data support the idea that HCV assembly occurs in concert with budding at the ER membrane. Furthermore, capsid-containing particles did not accumulate in the absence of SP-catalyzed cleavage, suggesting the quality of newly formed viral particles is controlled before secretion.
Conflict of interest statement
Figures




Similar articles
-
Signal peptide peptidase-catalyzed cleavage of hepatitis C virus core protein is dispensable for virus budding but destabilizes the viral capsid.J Biol Chem. 2006 Sep 22;281(38):27679-92. doi: 10.1074/jbc.M602587200. Epub 2006 Jul 18. J Biol Chem. 2006. PMID: 16849324
-
Sequential processing of hepatitis C virus core protein by host cell signal peptidase and signal peptide peptidase: a reassessment.J Viral Hepat. 2009 Oct;16(10):705-15. doi: 10.1111/j.1365-2893.2009.01118.x. Epub 2009 Mar 5. J Viral Hepat. 2009. PMID: 19281487
-
Core protein cleavage by signal peptide peptidase is required for hepatitis C virus-like particle assembly.J Gen Virol. 2006 Apr;87(Pt 4):855-860. doi: 10.1099/vir.0.81664-0. J Gen Virol. 2006. PMID: 16528035 Free PMC article.
-
Ultrastructural and biochemical basis for hepatitis C virus morphogenesis.Virus Genes. 2017 Apr;53(2):151-164. doi: 10.1007/s11262-017-1426-2. Epub 2017 Feb 23. Virus Genes. 2017. PMID: 28233195 Review.
-
Hepatitis C virus core protein, lipid droplets and steatosis.J Viral Hepat. 2008 Mar;15(3):157-64. doi: 10.1111/j.1365-2893.2007.00953.x. Epub 2007 Dec 17. J Viral Hepat. 2008. PMID: 18086178 Review.
Cited by
-
Murine astrotactins 1 and 2 have a similar membrane topology and mature via endoproteolytic cleavage catalyzed by a signal peptidase.J Biol Chem. 2019 Mar 22;294(12):4538-4545. doi: 10.1074/jbc.RA118.007093. Epub 2019 Jan 29. J Biol Chem. 2019. PMID: 30696770 Free PMC article.
-
The Transgene Expression of the Immature Form of the HCV Core Protein (C191) and the LncRNA MEG3 Increases Apoptosis in HepG2 Cells.Curr Issues Mol Biol. 2022 Aug 12;44(8):3632-3647. doi: 10.3390/cimb44080249. Curr Issues Mol Biol. 2022. PMID: 36005145 Free PMC article.
References
-
- McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. Journal of viral hepatitis. 2000;7(1):2–14. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

