Phase-plate cryo-EM structure of a class B GPCR-G-protein complex
- PMID: 28437792
- PMCID: PMC5832441
- DOI: 10.1038/nature22327
Phase-plate cryo-EM structure of a class B GPCR-G-protein complex
Abstract
Class B G-protein-coupled receptors are major targets for the treatment of chronic diseases, such as osteoporosis, diabetes and obesity. Here we report the structure of a full-length class B receptor, the calcitonin receptor, in complex with peptide ligand and heterotrimeric Gαsβγ protein determined by Volta phase-plate single-particle cryo-electron microscopy. The peptide agonist engages the receptor by binding to an extended hydrophobic pocket facilitated by the large outward movement of the extracellular ends of transmembrane helices 6 and 7. This conformation is accompanied by a 60° kink in helix 6 and a large outward movement of the intracellular end of this helix, opening the bundle to accommodate interactions with the α5-helix of Gαs. Also observed is an extended intracellular helix 8 that contributes to both receptor stability and functional G-protein coupling via an interaction with the Gβ subunit. This structure provides a new framework for understanding G-protein-coupled receptor function.
Conflict of interest statement
The authors declare no competing financial interests for this manuscript.
Figures

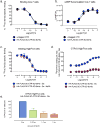
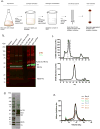
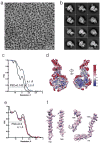











Comment in
-
Structural biology: Signalling under the microscope.Nature. 2017 Jun 1;546(7656):36-37. doi: 10.1038/nature22491. Epub 2017 May 3. Nature. 2017. PMID: 28467826 No abstract available.
References
-
- Hollenstein K, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. - PubMed

