A universal genome sequencing method for rotavirus A from human fecal samples which identifies segment reassortment and multi-genotype mixed infection
- PMID: 28438140
- PMCID: PMC5404283
- DOI: 10.1186/s12864-017-3714-6
A universal genome sequencing method for rotavirus A from human fecal samples which identifies segment reassortment and multi-genotype mixed infection
Abstract
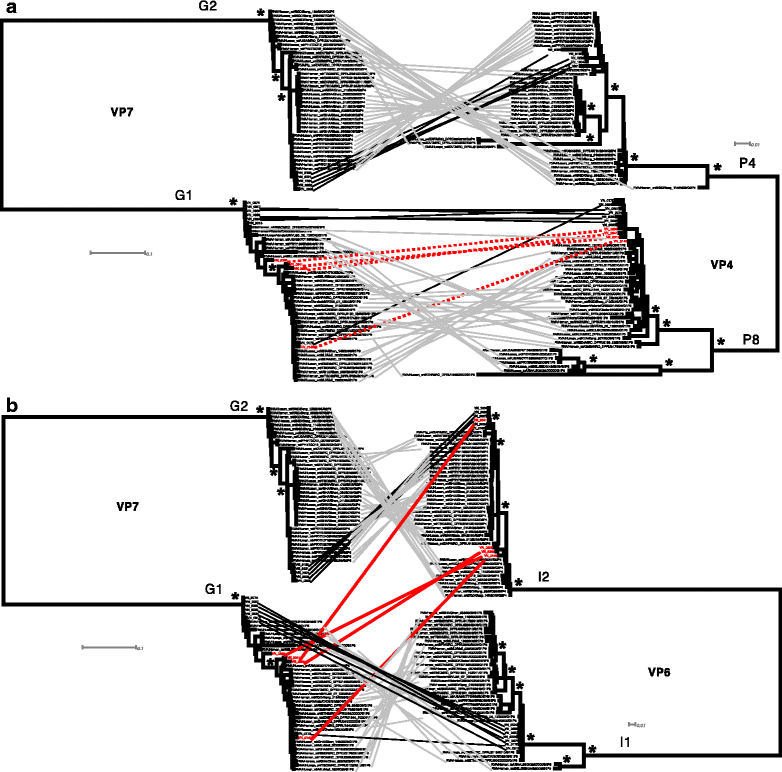
Background: Genomic characterization of rotavirus (RoV) has not been adopted at large-scale due to the complexity of obtaining sequences for all 11 segments, particularly when feces are used as starting material.
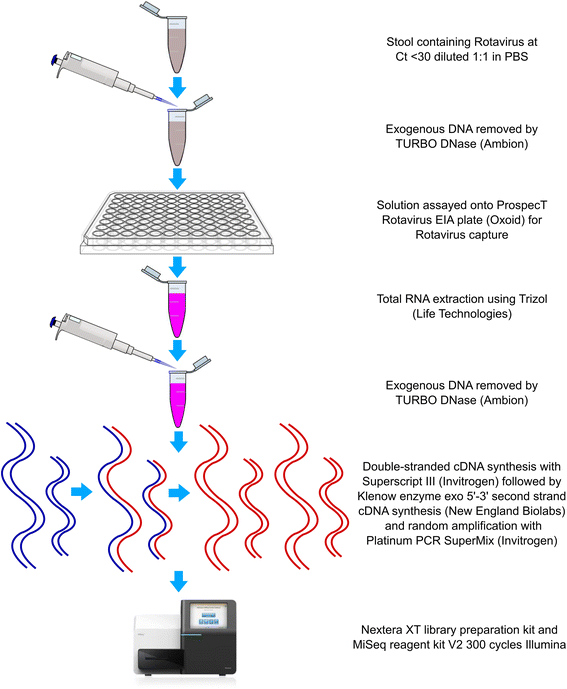
Methods: To overcome these limitations, we developed a novel RoV capture and genome sequencing method combining commercial enzyme immunoassay plates and a set of routinely used reagents.
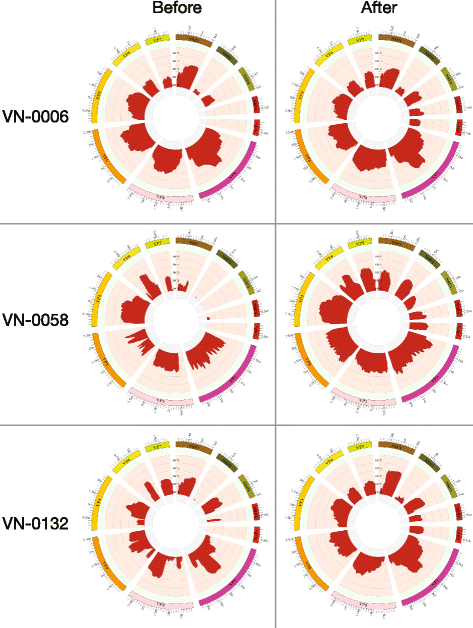
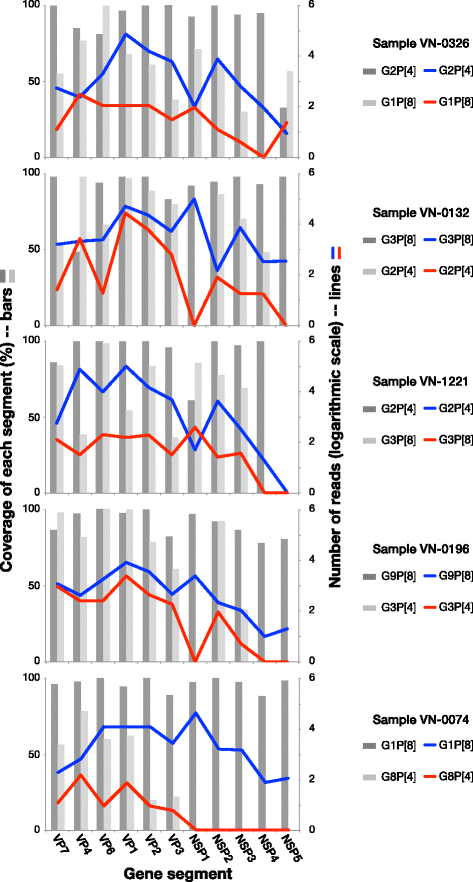
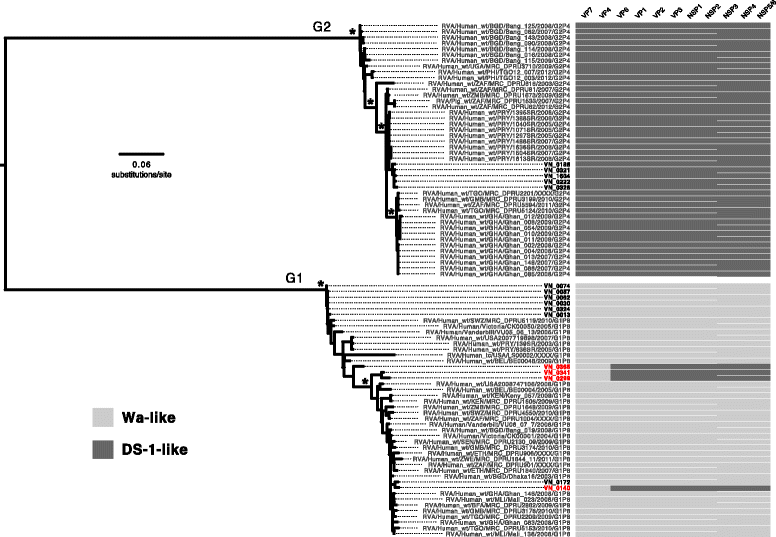
Results: Our approach had a 100% success rate, producing >90% genome coverage for diverse RoV present in fecal samples (Ct < 30).
Conclusions: This method provides a novel, reproducible and comparatively simple approach for genomic RoV characterization and could be scaled-up for use in global RoV surveillance systems.
Trial registration (prospectively registered): Current Controlled Trials ISRCTN88101063 . Date of registration: 14/06/2012.
Keywords: Antibody capture; Co-infection; Genome sequencing; Genomics; Phylogenetics; Reassortment; Rotavirus A.
Figures
References
-
- Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. - DOI - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

