PathOS: a decision support system for reporting high throughput sequencing of cancers in clinical diagnostic laboratories
- PMID: 28438193
- PMCID: PMC5404673
- DOI: 10.1186/s13073-017-0427-z
PathOS: a decision support system for reporting high throughput sequencing of cancers in clinical diagnostic laboratories
Abstract
Background: The increasing affordability of DNA sequencing has allowed it to be widely deployed in pathology laboratories. However, this has exposed many issues with the analysis and reporting of variants for clinical diagnostic use. Implementing a high-throughput sequencing (NGS) clinical reporting system requires a diverse combination of capabilities, statistical methods to identify variants, global variant databases, a validated bioinformatics pipeline, an auditable laboratory workflow, reproducible clinical assays and quality control monitoring throughout. These capabilities must be packaged in software that integrates the disparate components into a useable system.
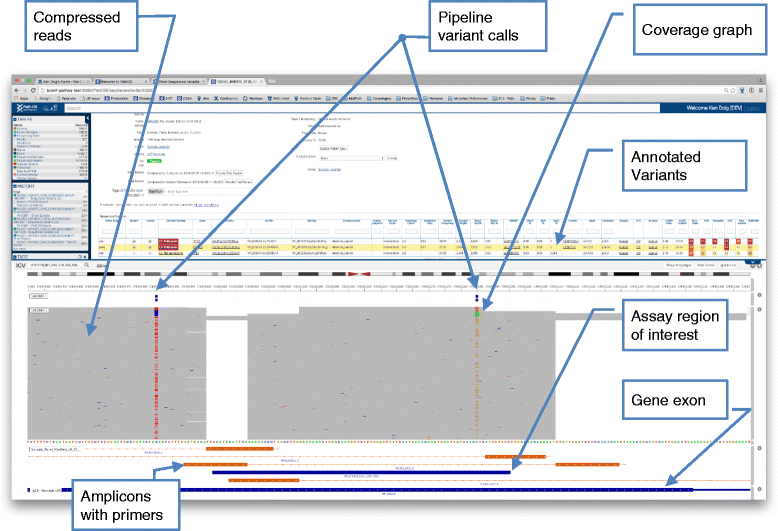
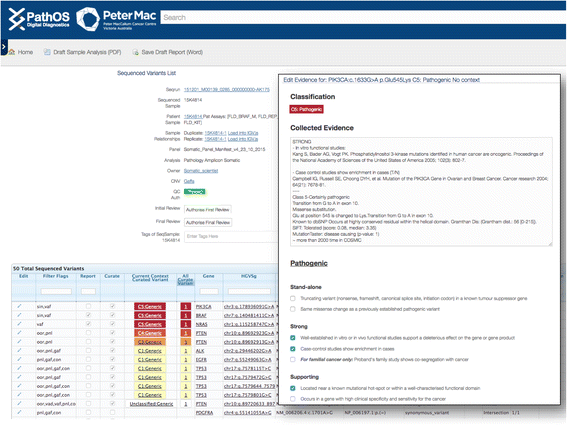
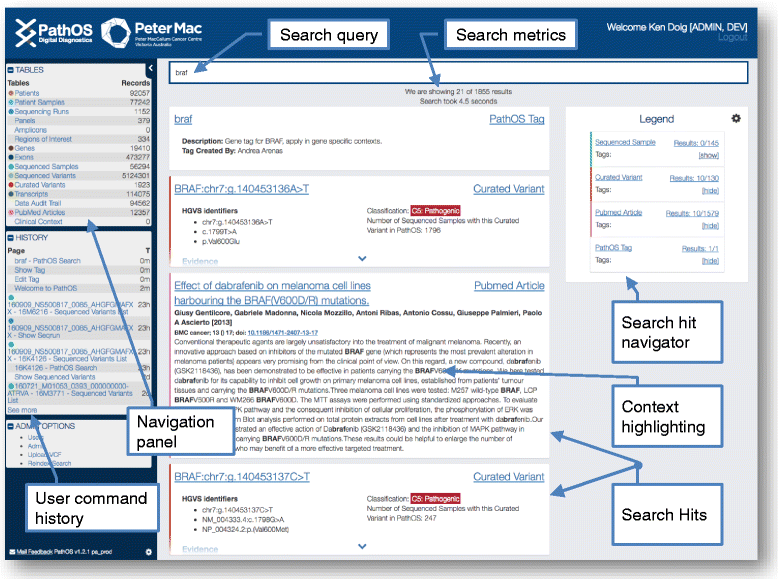
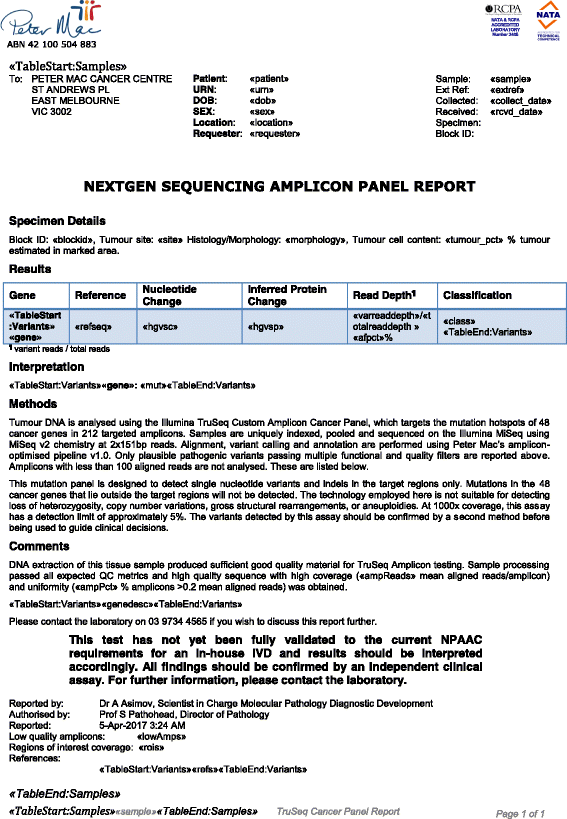
Results: To meet these needs, we developed a web-based application, PathOS, which takes variant data from a patient sample through to a clinical report. PathOS has been used operationally in the Peter MacCallum Cancer Centre for two years for the analysis, curation and reporting of genetic tests for cancer patients, as well as the curation of large-scale research studies. PathOS has also been deployed in cloud environments allowing multiple institutions to use separate, secure and customisable instances of the system. Increasingly, the bottleneck of variant curation is limiting the adoption of clinical sequencing for molecular diagnostics. PathOS is focused on providing clinical variant curators and pathology laboratories with a decision support system needed for personalised medicine. While the genesis of PathOS has been within cancer molecular diagnostics, the system is applicable to NGS clinical reporting generally.
Conclusions: The widespread availability of genomic sequencers has highlighted the limited availability of software to support clinical decision-making in molecular pathology. PathOS is a system that has been developed and refined in a hospital laboratory context to meet the needs of clinical diagnostics. The software is available as a set of Docker images and source code at https://github.com/PapenfussLab/PathOS .
Figures




References
-
- NATA. http://www.nata.com.au. Accessed 19 Apr 2017.
-
- Interactive_Biosoftware. Alamut Visual. 2016. http://www.interactive-biosoftware.com/alamut-visual/. Accessed 19 Apr 2017.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

