Distinct cis-acting regions control six6 expression during eye field and optic cup stages of eye formation
- PMID: 28438336
- PMCID: PMC5500183
- DOI: 10.1016/j.ydbio.2017.04.003
Distinct cis-acting regions control six6 expression during eye field and optic cup stages of eye formation
Abstract
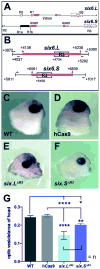
The eye field transcription factor, Six6, is essential for both the early (specification and proliferative growth) phase of eye formation, as well as for normal retinal progenitor cell differentiation. While genomic regions driving six6 optic cup expression have been described, the sequences controlling eye field and optic vesicle expression are unknown. Two evolutionary conserved regions 5' and a third 3' to the six6 coding region were identified, and together they faithfully replicate the endogenous X. laevis six6 expression pattern. Transgenic lines were generated and used to determine the onset and expression patterns controlled by the regulatory regions. The conserved 3' region was necessary and sufficient for eye field and optic vesicle expression. In contrast, the two conserved enhancer regions located 5' of the coding sequence were required together for normal optic cup and mature retinal expression. Gain-of-function experiments indicate endogenous six6 and GFP expression in F1 transgenic embryos are similarly regulated in response to candidate trans-acting factors. Importantly, CRISPR/CAS9-mediated deletion of the 3' eye field/optic vesicle enhancer in X. laevis, resulted in a reduction in optic vesicle size. These results identify the cis-acting regions, demonstrate the modular nature of the elements controlling early versus late retinal expression, and identify potential regulators of six6 expression during the early stages of eye formation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Co-accumulation of cis-regulatory and coding mutations during the pseudogenization of the Xenopus laevis homoeologs six6.L and six6.S.Dev Biol. 2017 Jul 1;427(1):84-92. doi: 10.1016/j.ydbio.2017.05.004. Epub 2017 May 10. Dev Biol. 2017. PMID: 28501477
-
Regulation of Pax6 expression is conserved between mice and flies.Development. 1999 Jan;126(2):383-95. doi: 10.1242/dev.126.2.383. Development. 1999. PMID: 9847251
-
Exploring the functions of nonclassical MHC class Ib genes in Xenopus laevis by the CRISPR/Cas9 system.Dev Biol. 2017 Jun 15;426(2):261-269. doi: 10.1016/j.ydbio.2016.05.023. Epub 2016 Jun 16. Dev Biol. 2017. PMID: 27318386 Free PMC article.
-
Structure, biological activity of the upstream regulatory sequence, and conserved domains of a middle molecular mass neurofilament gene of Xenopus laevis.Brain Res Mol Brain Res. 2000 Oct 20;82(1-2):35-51. doi: 10.1016/s0169-328x(00)00180-7. Brain Res Mol Brain Res. 2000. PMID: 11042356
-
Direct transcriptional regulation of Six6 is controlled by SoxB1 binding to a remote forebrain enhancer.Dev Biol. 2012 Jun 15;366(2):393-403. doi: 10.1016/j.ydbio.2012.04.023. Epub 2012 Apr 25. Dev Biol. 2012. PMID: 22561201 Free PMC article.
Cited by
-
Cell fate decisions, transcription factors and signaling during early retinal development.Prog Retin Eye Res. 2022 Nov;91:101093. doi: 10.1016/j.preteyeres.2022.101093. Epub 2022 Jul 8. Prog Retin Eye Res. 2022. PMID: 35817658 Free PMC article. Review.
-
The SIX Family of Transcription Factors: Common Themes Integrating Developmental and Cancer Biology.Front Cell Dev Biol. 2021 Aug 19;9:707854. doi: 10.3389/fcell.2021.707854. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34490256 Free PMC article. Review.
-
Temporal Transcriptomic Profiling of the Developing Xenopus laevis Eye.bioRxiv [Preprint]. 2024 Jul 22:2024.07.20.603187. doi: 10.1101/2024.07.20.603187. bioRxiv. 2024. Update in: Cells. 2024 Aug 21;13(16):1390. doi: 10.3390/cells13161390. PMID: 39091861 Free PMC article. Updated. Preprint.
-
Aberrant activity of NKL homeobox gene NKX3-2 in a T-ALL subset.PLoS One. 2018 May 10;13(5):e0197194. doi: 10.1371/journal.pone.0197194. eCollection 2018. PLoS One. 2018. PMID: 29746601 Free PMC article.
-
Optimization of CRISPR/Cas9-mediated gene disruption in Xenopus laevis using a phenotypic image analysis technique.Dev Growth Differ. 2022 May;64(4):219-225. doi: 10.1111/dgd.12778. Epub 2022 Apr 12. Dev Growth Differ. 2022. PMID: 35338712 Free PMC article.
References
-
- Aijaz S, Allen J, Tregidgo R, van Heyningen V, Hanson I, Clark BJ. Expression analysis of SIX3 and SIX6 in human tissues reveals differences in expression and a novel correlation between the expression of SIX3 and the genes encoding isocitrate dehyhrogenase and cadherin 18. Genomics. 2005;86:86–99. - PubMed
-
- Aijaz S, Clark BJ, Williamson K, van Heyningen V, Morrison D, Fitzpatrick D, Collin R, Ragge N, Christoforou A, Brown A, Hanson I. Absence of SIX6 mutations in microphthalmia, anophthalmia, and coloboma. Invest Ophthalmol Vis Sci. 2004;45:3871–3876. - PubMed
-
- Aldahmesh MA, Khan AO, Hijazi H, Alkuraya FS. Homozygous truncation of SIX6 causes complex microphthalmia in humans. Clin Genet. 2013;84:198–199. - PubMed
-
- Bar-Yosef U, Abuelaish I, Harel T, Hendler N, Ofir R, Birk OS. CHX10 mutations cause non-syndromic microphthalmia/anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

