Emerging Affinity-Based Proteomic Technologies for Large-Scale Plasma Profiling in Cardiovascular Disease
- PMID: 28438806
- PMCID: PMC5555416
- DOI: 10.1161/CIRCULATIONAHA.116.025446
Emerging Affinity-Based Proteomic Technologies for Large-Scale Plasma Profiling in Cardiovascular Disease
Abstract
Plasma biomarkers that reflect molecular states of the cardiovascular system are central for clinical decision making. Routinely used plasma biomarkers include troponins, natriuretic peptides, and lipoprotein particles, yet interrogate only a modest subset of pathways relevant to cardiovascular disease. Systematic profiling of a larger portion of circulating plasma proteins (the plasma proteome) will provide opportunities for unbiased discovery of novel markers to improve diagnostic or predictive accuracy. In addition, proteomic profiling may inform pathophysiological understanding and point to novel therapeutic targets. Obstacles for comprehensive proteomic profiling include the immense size and structural heterogeneity of the proteome, and the broad range of abundance levels, as well. Proteome-wide, untargeted profiling can be performed in tissues and cells with tandem mass spectrometry. However, applications to plasma are limited by the need for complex preanalytical sample preparation stages limiting sample throughput. Multiplexing of targeted methods based on capture and detection of specific proteins are therefore receiving increasing attention in plasma proteomics. Immunoaffinity assays are the workhorse for measuring individual proteins but have been limited for proteomic applications by long development times, cross-reactivity preventing multiplexing, specificity issues, and incomplete sensitivity to detect proteins in the lower range of the abundance spectrum (below picograms per milliliter). Emerging technologies to address these issues include nucleotide-labeled immunoassays and aptamer reagents that can be automated for efficient multiplexing of thousands of proteins at high sample throughput, coupling of affinity capture methods to mass spectrometry for improved specificity, and ultrasensitive detection systems to measure low-abundance proteins. In addition, proteomics can now be integrated with modern genomics tools to comprehensively relate proteomic profiles to genetic variants, which may both influence binding of affinity reagents and serve to validate the target specificity of affinity assays. The application of deep quantitative proteomic profiling to large cohorts has thus become increasingly feasible with emerging affinity methods. The aims of this article are to provide the broad readership of Circulation with a timely overview of emerging methods for affinity proteomics and recent progress in cardiovascular medicine based on such methods.
Keywords: biomarkers; cardiovascular diseases; epidemiology; plasma; proteomics.
© 2017 American Heart Association, Inc.
Figures
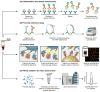
References
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. - PMC - PubMed
-
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. - PubMed
-
- Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. A functional annotation of subproteomes in human plasma. Proteomics. 2005;5:3506–19. - PubMed
-
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular & cellular proteomics: MCP. 2002;1:845–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

