Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M Integrated in the Chromosome
- PMID: 28438939
- PMCID: PMC5487645
- DOI: 10.1128/AAC.00076-17
Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M Integrated in the Chromosome
Abstract
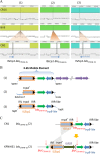
The extended-spectrum-β-lactamase (ESBL)- and Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae represent serious and urgent threats to public health. In a retrospective study of multidrug-resistant K. pneumoniae, we identified three clinical isolates, CN1, CR14, and NY9, carrying both blaCTX-M and blaKPC genes. The complete genomes of these three K. pneumoniae isolates were de novo assembled by using both short- and long-read whole-genome sequencing. In CR14 and NY9, blaCTX-M and blaKPC were carried on two different plasmids. In contrast, CN1 had one copy of blaKPC-2 and three copies of blaCTX-M-15 integrated in the chromosome, for which the blaCTX-M-15 genes were linked to an insertion sequence, ISEcp1, whereas the blaKPC-2 gene was in the context of a Tn4401a transposition unit conjugated with a PsP3-like prophage. Intriguingly, downstream of the Tn4401a-blaKPC-2-prophage genomic island, CN1 also carried a clustered regularly interspaced short palindromic repeat (CRISPR)-cas array with four spacers targeting a variety of K. pneumoniae plasmids harboring antimicrobial resistance genes. Comparative genomic analysis revealed that there were two subtypes of type I-E CRISPR-cas in K. pneumoniae strains and suggested that the evolving CRISPR-cas, with its acquired novel spacer, induced the mobilization of antimicrobial resistance genes from plasmids into the chromosome. The integration and dissemination of multiple copies of blaCTX-M and blaKPC from plasmids to chromosome depicts the complex pandemic scenario of multidrug-resistant K. pneumoniae Additionally, the implications from this study also raise concerns for the application of a CRISPR-cas strategy against antimicrobial resistance.
Keywords: CRISPR-Cas; CTX-M; Klebsiella pneumoniae; blaKPC; carbapenem-resistance; chromosomal beta-lactamases.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA.
-
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi:10.1128/AAC.00126-09. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

