Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair
- PMID: 28439008
- PMCID: PMC5441711
- DOI: 10.1073/pnas.1702561114
Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair
Abstract
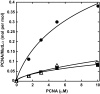
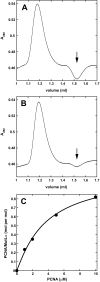
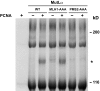
Eukaryotic MutLα (mammalian MLH1-PMS2 heterodimer; MLH1-PMS1 in yeast) functions in early steps of mismatch repair as a latent endonuclease that requires a mismatch, MutSα/β, and DNA-loaded proliferating cell nuclear antigen (PCNA) for activation. We show here that human PCNA and MutLα interact specifically but weakly in solution to form a complex of approximately 1:1 stoichiometry that depends on PCNA interaction with the C-terminal endonuclease domain of the MutLα PMS2 subunit. Amino acid substitution mutations within a PMS2 C-terminal 721QRLIAP motif attenuate or abolish human MutLα interaction with PCNA, as well as PCNA-dependent activation of MutLα endonuclease, PCNA- and DNA-dependent activation of MutLα ATPase, and MutLα function in in vitro mismatch repair. Amino acid substitution mutations within the corresponding yeast PMS1 motif (723QKLIIP) reduce or abolish mismatch repair in vivo. Coupling of a weak allele within this motif (723AKLIIP) with an exo1Δ null mutation, which individually confer only weak mutator phenotypes, inactivates mismatch repair in the yeast cell.
Keywords: DNA repair; MutLalpha; endonuclease; mismatch repair; proliferating cell nuclear antigen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease.J Biol Chem. 2007 Dec 21;282(51):37181-90. doi: 10.1074/jbc.M707617200. Epub 2007 Oct 19. J Biol Chem. 2007. PMID: 17951253 Free PMC article.
-
The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans.PLoS Biol. 2017 Apr 28;15(4):e2001164. doi: 10.1371/journal.pbio.2001164. eCollection 2017 Apr. PLoS Biol. 2017. PMID: 28453523 Free PMC article.
-
The unstructured linker of Mlh1 contains a motif required for endonuclease function which is mutated in cancers.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2212870119. doi: 10.1073/pnas.2212870119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215471 Free PMC article.
-
Endonuclease activities of MutLα and its homologs in DNA mismatch repair.DNA Repair (Amst). 2016 Feb;38:42-49. doi: 10.1016/j.dnarep.2015.11.023. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26719141 Free PMC article. Review.
-
Expanded roles for the MutL family of DNA mismatch repair proteins.Yeast. 2021 Jan;38(1):39-53. doi: 10.1002/yea.3512. Epub 2020 Jul 30. Yeast. 2021. PMID: 32652606 Free PMC article. Review.
Cited by
-
Mlh1 interacts with both Msh2 and Msh6 for recruitment during mismatch repair.DNA Repair (Amst). 2022 Nov;119:103405. doi: 10.1016/j.dnarep.2022.103405. Epub 2022 Sep 14. DNA Repair (Amst). 2022. PMID: 36122480 Free PMC article.
-
Intrinsically disordered regions regulate both catalytic and non-catalytic activities of the MutLα mismatch repair complex.Nucleic Acids Res. 2019 Feb 28;47(4):1823-1835. doi: 10.1093/nar/gky1244. Nucleic Acids Res. 2019. PMID: 30541127 Free PMC article.
-
A conserved motif in the disordered linker of human MLH1 is vital for DNA mismatch repair and its function is diminished by a cancer family mutation.Nucleic Acids Res. 2023 Jul 7;51(12):6307-6320. doi: 10.1093/nar/gkad418. Nucleic Acids Res. 2023. PMID: 37224528 Free PMC article.
-
Combining single-molecule and structural studies reveals protein and DNA conformations and assemblies that govern DNA mismatch repair.Curr Opin Struct Biol. 2024 Dec;89:102917. doi: 10.1016/j.sbi.2024.102917. Epub 2024 Sep 10. Curr Opin Struct Biol. 2024. PMID: 39260099 Review.
-
Functions of PMS2 and MLH1 important for regulation of divergent repeat-mediated deletions.bioRxiv [Preprint]. 2024 Aug 6:2024.08.05.606388. doi: 10.1101/2024.08.05.606388. bioRxiv. 2024. Update in: DNA Repair (Amst). 2025 Jan;145:103791. doi: 10.1016/j.dnarep.2024.103791. PMID: 39149360 Free PMC article. Updated. Preprint.
References
-
- Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. - PubMed
-
- Rowley PT. Inherited susceptibility to colorectal cancer. Annu Rev Med. 2005;56:539–554. - PubMed
-
- Jacinto FV, Esteller M. Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis. 2007;22:247–253. - PubMed
-
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

