CYCLOPS reveals human transcriptional rhythms in health and disease
- PMID: 28439010
- PMCID: PMC5441789
- DOI: 10.1073/pnas.1619320114
CYCLOPS reveals human transcriptional rhythms in health and disease
Abstract
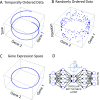
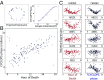
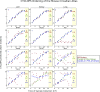
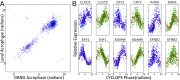
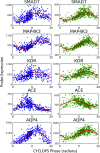
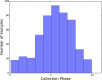
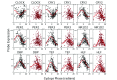
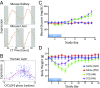
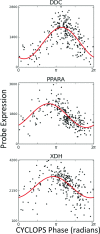
Circadian rhythms modulate many aspects of physiology. Knowledge of the molecular basis of these rhythms has exploded in the last 20 years. However, most of these data are from model organisms, and translation to clinical practice has been limited. Here, we present an approach to identify molecular rhythms in humans from thousands of unordered expression measurements. Our algorithm, cyclic ordering by periodic structure (CYCLOPS), uses evolutionary conservation and machine learning to identify elliptical structure in high-dimensional data. From this structure, CYCLOPS estimates the phase of each sample. We validated CYCLOPS using temporally ordered mouse and human data and demonstrated its consistency on human data from two independent research sites. We used this approach to identify rhythmic transcripts in human liver and lung, including hundreds of drug targets and disease genes. Importantly, for many genes, the circadian variation in expression exceeded variation from genetic and other environmental factors. We also analyzed hepatocellular carcinoma samples and show these solid tumors maintain circadian function but with aberrant output. Finally, to show how this method can catalyze medical translation, we show that dosage time can temporally segregate efficacy from dose-limiting toxicity of streptozocin, a chemotherapeutic drug. In sum, these data show the power of CYCLOPS and temporal reconstruction in bridging basic circadian research and clinical medicine.
Keywords: autoencoder; biological rhythms; circadian rhythms; gene expression; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Compass in the data ocean: Toward chronotherapy.Proc Natl Acad Sci U S A. 2017 May 16;114(20):5069-5071. doi: 10.1073/pnas.1705326114. Epub 2017 May 9. Proc Natl Acad Sci U S A. 2017. PMID: 28487482 Free PMC article. No abstract available.
-
An "eye" for rhythm.Sci Transl Med. 2017 May 17;9(390):eaan4288. doi: 10.1126/scitranslmed.aan4288. Sci Transl Med. 2017. PMID: 28515338
References
-
- Straub RH, Cutolo M. Circadian rhythms in rheumatoid arthritis: Implications for pathophysiology and therapeutic management. Arthritis Rheum. 2007;56:399–408. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

