Intriguing Interaction of Bacteriophage-Host Association: An Understanding in the Era of Omics
- PMID: 28439260
- PMCID: PMC5383658
- DOI: 10.3389/fmicb.2017.00559
Intriguing Interaction of Bacteriophage-Host Association: An Understanding in the Era of Omics
Abstract
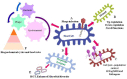
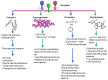
Innovations in next-generation sequencing technology have introduced new avenues in microbial studies through "omics" approaches. This technology has considerably augmented the knowledge of the microbial world without isolation prior to their identification. With an enormous volume of bacterial "omics" data, considerable attempts have been recently invested to improve an insight into virosphere. The interplay between bacteriophages and their host has created a significant influence on the biogeochemical cycles, microbial diversity, and bacterial population regulation. This review highlights various concepts such as genomics, transcriptomics, proteomics, and metabolomics to infer the phylogenetic affiliation and function of bacteriophages and their impact on diverse microbial communities. Omics technologies illuminate the role of bacteriophage in an environment, the influences of phage proteins on the bacterial host and provide information about the genes important for interaction with bacteria. These investigations will reveal some of bio-molecules and biomarkers of the novel phage which demand to be unveiled.
Keywords: bacteriophage; genomics; next-generation sequencing; transcriptomics.
Figures

Similar articles
-
Alzheimer's disease in the omics era.Clin Biochem. 2018 Sep;59:9-16. doi: 10.1016/j.clinbiochem.2018.06.011. Epub 2018 Jun 18. Clin Biochem. 2018. PMID: 29920246 Review.
-
Progress and promise of omics for predicting the impacts of climate change on harmful algal blooms.Harmful Algae. 2020 Jan;91:101587. doi: 10.1016/j.hal.2019.03.005. Epub 2019 Jun 8. Harmful Algae. 2020. PMID: 32057337 Review.
-
A role for ecophysiology in the 'omics' era.Plant J. 2018 Oct;96(2):251-259. doi: 10.1111/tpj.14059. Epub 2018 Sep 19. Plant J. 2018. PMID: 30091802 Review.
-
Next-generation technologies for multiomics approaches including interactome sequencing.Biomed Res Int. 2015;2015:104209. doi: 10.1155/2015/104209. Epub 2015 Jan 12. Biomed Res Int. 2015. PMID: 25649523 Free PMC article. Review.
-
Next-Generation Sequencing: The Translational Medicine Approach from "Bench to Bedside to Population".Medicines (Basel). 2016 Jun 2;3(2):14. doi: 10.3390/medicines3020014. Medicines (Basel). 2016. PMID: 28930123 Free PMC article. Review.
Cited by
-
Transcriptional Landscapes of Herelleviridae Bacteriophages and Staphylococcus aureus during Phage Infection: An Overview.Viruses. 2023 Jun 23;15(7):1427. doi: 10.3390/v15071427. Viruses. 2023. PMID: 37515114 Free PMC article. Review.
-
Analysis of Microbial Functions in the Rhizosphere Using a Metabolic-Network Based Framework for Metagenomics Interpretation.Front Microbiol. 2017 Aug 23;8:1606. doi: 10.3389/fmicb.2017.01606. eCollection 2017. Front Microbiol. 2017. PMID: 28878756 Free PMC article.
-
Interlaboratory comparison of Pseudomonas aeruginosa phage susceptibility testing.J Clin Microbiol. 2023 Dec 19;61(12):e0061423. doi: 10.1128/jcm.00614-23. Epub 2023 Nov 14. J Clin Microbiol. 2023. PMID: 37962552 Free PMC article.
-
Development of ONT-cappable-seq to unravel the transcriptional landscape of Pseudomonas phages.Comput Struct Biotechnol J. 2022 May 23;20:2624-2638. doi: 10.1016/j.csbj.2022.05.034. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35685363 Free PMC article.
-
Adaptation of metal and antibiotic resistant traits in novel β-Proteobacterium Achromobacter xylosoxidans BHW-15.PeerJ. 2019 Mar 13;7:e6537. doi: 10.7717/peerj.6537. eCollection 2019. PeerJ. 2019. PMID: 30886770 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

