Microbial Diversity of Browning Peninsula, Eastern Antarctica Revealed Using Molecular and Cultivation Methods
- PMID: 28439263
- PMCID: PMC5383709
- DOI: 10.3389/fmicb.2017.00591
Microbial Diversity of Browning Peninsula, Eastern Antarctica Revealed Using Molecular and Cultivation Methods
Abstract
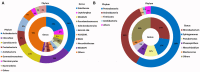
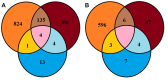
Browning Peninsula is an ice-free polar desert situated in the Windmill Islands, Eastern Antarctica. The entire site is described as a barren landscape, comprised of frost boils with soils dominated by microbial life. In this study, we explored the microbial diversity and edaphic drivers of community structure across this site using traditional cultivation methods, a novel approach the soil substrate membrane system (SSMS), and culture-independent 454-tag pyrosequencing. The measured soil environmental and microphysical factors of chlorine, phosphate, aspect and elevation were found to be significant drivers of the bacterial community, while none of the soil parameters analyzed were significantly correlated to the fungal community. Overall, Browning Peninsula soil harbored a distinctive microbial community in comparison to other Antarctic soils comprised of a unique bacterial diversity and extremely limited fungal diversity. Tag pyrosequencing data revealed the bacterial community to be dominated by Actinobacteria (36%), followed by Chloroflexi (18%), Cyanobacteria (14%), and Proteobacteria (10%). For fungi, Ascomycota (97%) dominated the soil microbiome, followed by Basidiomycota. As expected the diversity recovered from culture-based techniques was lower than that detected using tag sequencing. However, in the SSMS enrichments, that mimic the natural conditions for cultivating oligophilic "k-selected" bacteria, a larger proportion of rare bacterial taxa (15%), such as Blastococcus, Devosia, Herbaspirillum, Propionibacterium and Methylocella and fungal (11%) taxa, such as Nigrospora, Exophiala, Hortaea, and Penidiella were recovered at the genus level. At phylum level, a comparison of OTU's showed that the SSMS shared 21% of Acidobacteria, 11% of Actinobacteria and 10% of Proteobacteria OTU's with soil. For fungi, the shared OTUs was 4% (Basidiomycota) and <0.5% (Ascomycota). This was the first known attempt to culture microfungi using the SSMS which resulted in an increase in diversity from 14 to 57 microfungi OTUs compared to standard cultivation. Furthermore, the SSMS offers the opportunity to retrieve a greater diversity of bacterial and fungal taxa for future exploitation.
Keywords: antarctic soil; bacterial diversity; cultivation; frost boils; fungal diversity.
Figures


Similar articles
-
Yucatán in black and red: Linking edaphic analysis and pyrosequencing-based assessment of bacterial and fungal community structures in the two main kinds of soil of Yucatán State.Microbiol Res. 2016 Jul-Aug;188-189:23-33. doi: 10.1016/j.micres.2016.04.007. Epub 2016 Apr 22. Microbiol Res. 2016. PMID: 27296959
-
Fungi in the Antarctic Cryosphere: Using DNA Metabarcoding to Reveal Fungal Diversity in Glacial Ice from the Antarctic Peninsula Region.Microb Ecol. 2022 Apr;83(3):647-657. doi: 10.1007/s00248-021-01792-x. Epub 2021 Jul 6. Microb Ecol. 2022. PMID: 34228196
-
Novel Culturing Techniques Select for Heterotrophs and Hydrocarbon Degraders in a Subantarctic Soil.Sci Rep. 2016 Nov 9;6:36724. doi: 10.1038/srep36724. Sci Rep. 2016. PMID: 27827405 Free PMC article.
-
Uncovering the Uncultivated Majority in Antarctic Soils: Toward a Synergistic Approach.Front Microbiol. 2019 Feb 15;10:242. doi: 10.3389/fmicb.2019.00242. eCollection 2019. Front Microbiol. 2019. PMID: 30828325 Free PMC article. Review.
-
Are uncultivated bacteria really uncultivable?Microbes Environ. 2012;27(4):356-66. doi: 10.1264/jsme2.me12092. Epub 2012 Oct 10. Microbes Environ. 2012. PMID: 23059723 Free PMC article. Review.
Cited by
-
Active antibiotic resistome in soils unraveled by single-cell isotope probing and targeted metagenomics.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2201473119. doi: 10.1073/pnas.2201473119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161886 Free PMC article.
-
Microbial Community Composition of the Antarctic Ecosystems: Review of the Bacteria, Fungi, and Archaea Identified through an NGS-Based Metagenomics Approach.Life (Basel). 2022 Jun 18;12(6):916. doi: 10.3390/life12060916. Life (Basel). 2022. PMID: 35743947 Free PMC article. Review.
-
Actinobacteria and Cyanobacteria Diversity in Terrestrial Antarctic Microenvironments Evaluated by Culture-Dependent and Independent Methods.Front Microbiol. 2019 May 31;10:1018. doi: 10.3389/fmicb.2019.01018. eCollection 2019. Front Microbiol. 2019. PMID: 31214128 Free PMC article.
-
Soil substrate culturing approaches recover diverse members of Actinomycetota from desert soils of Herring Island, East Antarctica.Extremophiles. 2022 Jul 13;26(2):24. doi: 10.1007/s00792-022-01271-2. Extremophiles. 2022. PMID: 35829965 Free PMC article.
-
Development of PCR-based molecular marker for screening of disease-suppressive composts against Fusarium wilt of tomato (Solanum lycopersicum L.).3 Biotech. 2018 Jul;8(7):306. doi: 10.1007/s13205-018-1331-y. Epub 2018 Jul 7. 3 Biotech. 2018. PMID: 30002996 Free PMC article.
References
-
- Anderson M. J., Gorley M. J., Clarke K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth: PRIMER-E.
-
- Arenz B. E., Blanchette R. A. (2011). Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region and McMurdo Dry Valleys. Soil Biol. Biochem. 43, 308–315. 10.1016/j.soilbio.2010.10.016 - DOI
-
- Arenz B. E., Held B. W., Jurgens J. A., Farrell R. L., Blanchette R. A. (2006). Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 38, 3057–3064. 10.1016/j.soilbio.2006.01.016 - DOI
-
- Azmi O. R., Seppelt R. D. (1998). The broad-scale distribution of microfungi in the Windmill Islands region, continental Antarctica. Polar Biol. 19, 92–100. 10.1007/s003000050219 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

