Exploiting induced pluripotent stem cell-derived macrophages to unravel host factors influencing Chlamydia trachomatis pathogenesis
- PMID: 28440293
- PMCID: PMC5414054
- DOI: 10.1038/ncomms15013
Exploiting induced pluripotent stem cell-derived macrophages to unravel host factors influencing Chlamydia trachomatis pathogenesis
Abstract
Chlamydia trachomatis remains a leading cause of bacterial sexually transmitted infections and preventable blindness worldwide. There are, however, limited in vitro models to study the role of host genetics in the response of macrophages to this obligate human pathogen. Here, we describe an approach using macrophages derived from human induced pluripotent stem cells (iPSdMs) to study macrophage-Chlamydia interactions in vitro. We show that iPSdMs support the full infectious life cycle of C. trachomatis in a manner that mimics the infection of human blood-derived macrophages. Transcriptomic and proteomic profiling of the macrophage response to chlamydial infection highlighted the role of the type I interferon and interleukin 10-mediated responses. Using CRISPR/Cas9 technology, we generated biallelic knockout mutations in host genes encoding IRF5 and IL-10RA in iPSCs, and confirmed their roles in limiting chlamydial infection in macrophages. This model can potentially be extended to other pathogens and tissue systems to advance our understanding of host-pathogen interactions and the role of human genetics in influencing the outcome of infections.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

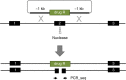
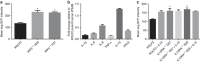

Similar articles
-
Transcriptional profiling of human epithelial cells infected with plasmid-bearing and plasmid-deficient Chlamydia trachomatis.Infect Immun. 2015 Feb;83(2):534-43. doi: 10.1128/IAI.02764-14. Epub 2014 Nov 17. Infect Immun. 2015. PMID: 25404022 Free PMC article.
-
Early Transcriptional Landscapes of Chlamydia trachomatis-Infected Epithelial Cells at Single Cell Resolution.Front Cell Infect Microbiol. 2019 Nov 19;9:392. doi: 10.3389/fcimb.2019.00392. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31803632 Free PMC article.
-
Extrusions are phagocytosed and promote Chlamydia survival within macrophages.Cell Microbiol. 2017 Apr;19(4). doi: 10.1111/cmi.12683. Epub 2016 Nov 21. Cell Microbiol. 2017. PMID: 27739160
-
Chlamydia cell biology and pathogenesis.Nat Rev Microbiol. 2016 Jun;14(6):385-400. doi: 10.1038/nrmicro.2016.30. Epub 2016 Apr 25. Nat Rev Microbiol. 2016. PMID: 27108705 Free PMC article. Review.
-
Pathogenesis of fallopian tube damage caused by Chlamydia trachomatis infections.Contraception. 2015 Aug;92(2):108-15. doi: 10.1016/j.contraception.2015.01.004. Epub 2015 Jan 13. Contraception. 2015. PMID: 25592078 Review.
Cited by
-
Induced Pluripotent Stem Cell-Derived Monocytes/Macrophages in Autoinflammatory Diseases.Front Immunol. 2022 May 6;13:870535. doi: 10.3389/fimmu.2022.870535. eCollection 2022. Front Immunol. 2022. PMID: 35603217 Free PMC article. Review.
-
Clear Victory for Chlamydia: The Subversion of Host Innate Immunity.Front Microbiol. 2019 Jul 3;10:1412. doi: 10.3389/fmicb.2019.01412. eCollection 2019. Front Microbiol. 2019. PMID: 31333596 Free PMC article. Review.
-
Human Induced Pluripotent Stem Cell-Derived Macrophages for Unraveling Human Macrophage Biology.Arterioscler Thromb Vasc Biol. 2017 Nov;37(11):2000-2006. doi: 10.1161/ATVBAHA.117.309195. Epub 2017 Oct 5. Arterioscler Thromb Vasc Biol. 2017. PMID: 28982665 Free PMC article. Review.
-
Development of innate immune cells from human pluripotent stem cells.Exp Hematol. 2019 Mar;71:13-23. doi: 10.1016/j.exphem.2018.12.005. Epub 2019 Jan 4. Exp Hematol. 2019. PMID: 30611869 Free PMC article. Review.
-
Novel stem cell technologies are powerful tools to understand the impact of human factors on Plasmodium falciparum malaria.Front Cell Infect Microbiol. 2023 Dec 19;13:1287355. doi: 10.3389/fcimb.2023.1287355. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38173794 Free PMC article.
References
-
- Hafner L., Beagley K. & Timms P. Chlamydia trachomatis infection: host immune responses and potential vaccines. Mucosal Immunol. 1, 116–130 (2008). - PubMed
-
- Beutler A. M., Schumacher H. R. Jr, Whittum-Hudson J. A., Salameh W. A. & Hudson A. P. Case report: in situ hybridization for dection of inapparent infection with Chlamydia trachomatis in synovial tissue of a patient with Reiter's syndrome. Am. J. Med. Sci. 310, 206–213 (1995). - PubMed
-
- Gerard H. C., Branigan P. J., Schumacher H. R. Jr & Hudson A. P. Synovial Chlamydia trachomatis in patients with reactive arthritis/Reiter's syndrome are viable but show aberrant gene expression. J. Rheumatol. 25, 734–742 (1998). - PubMed
-
- Nanagara R., Li F., Beutler A., Hudson A. & Schumacher H. R. Jr Alteration of Chlamydia trachomatis biologic behaviour in synovial membranes. Suppression of surface antigen production in reactive arthritis and Reiter's syndrome. Arthritis Rheum. 38, 1410–1417 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

