Significant variation between SNP-based HLA imputations in diverse populations: the last mile is the hardest
- PMID: 28440342
- PMCID: PMC5656547
- DOI: 10.1038/tpj.2017.7
Significant variation between SNP-based HLA imputations in diverse populations: the last mile is the hardest
Abstract
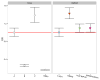
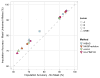
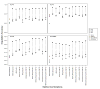
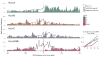
Four single nucleotide polymorphism (SNP)-based human leukocyte antigen (HLA) imputation methods (e-HLA, HIBAG, HLA*IMP:02 and MAGPrediction) were trained using 1000 Genomes SNP and HLA genotypes and assessed for their ability to accurately impute molecular HLA-A, -B, -C and -DRB1 genotypes in the Human Genome Diversity Project cell panel. Imputation concordance was high (>89%) across all methods for both HLA-A and HLA-C, but HLA-B and HLA-DRB1 proved generally difficult to impute. Overall, <27.8% of subjects were correctly imputed for all HLA loci by any method. Concordance across all loci was not enhanced via the application of confidence thresholds; reliance on confidence scores across methods only led to noticeable improvement (+3.2%) for HLA-DRB1. As the HLA complex is highly relevant to the study of human health and disease, a standardized assessment of SNP-based HLA imputation methods is crucial for advancing genomic research. Considerable room remains for the improvement of HLA-B and especially HLA-DRB1 imputation methods, and no imputation method is as accurate as molecular genotyping. The application of large, ancestrally diverse HLA and SNP reference data sets and multiple imputation methods has the potential to make SNP-based HLA imputation methods a tractable option for determining HLA genotypes.
Conflict of interest statement
SL is a founder and partner in Peptide Groove LLP. All other authors of this manuscript declare no competing financial interests.
Figures


References
-
- Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359(9308):727–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

