Matrix Metalloproteinase Gene Activation Resulting from Disordred Epigenetic Mechanisms in Rheumatoid Arthritis
- PMID: 28441353
- PMCID: PMC5454818
- DOI: 10.3390/ijms18050905
Matrix Metalloproteinase Gene Activation Resulting from Disordred Epigenetic Mechanisms in Rheumatoid Arthritis
Abstract
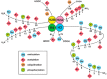
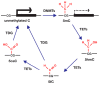
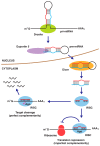
Matrix metalloproteinases (MMPs) are implicated in the degradation of extracellular matrix (ECM). Rheumatoid arthritis (RA) synovial fibroblasts (SFs) produce matrix-degrading enzymes, including MMPs, which facilitate cartilage destruction in the affected joints in RA. Epigenetic mechanisms contribute to change in the chromatin state, resulting in an alteration of gene transcription. Recently, MMP gene activation has been shown to be caused in RASFs by the dysregulation of epigenetic changes, such as histone modifications, DNA methylation, and microRNA (miRNA) signaling. In this paper, we review the role of MMPs in the pathogenesis of RA as well as the disordered epigenetic mechanisms regulating MMP gene activation in RASFs.
Keywords: epigenetics; gene transcription; matrix metalloproteinase; rheumatoid arthritis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

