Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study
- PMID: 28441386
- PMCID: PMC5404754
- DOI: 10.1371/journal.pmed.1002286
Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study
Abstract
Background: Graft-derived cell-free DNA (GcfDNA), which is released into the blood stream by necrotic and apoptotic cells, is a promising noninvasive organ integrity biomarker. In liver transplantation (LTx), neither conventional liver function tests (LTFs) nor immunosuppressive drug monitoring are very effective for rejection monitoring. We therefore hypothesized that the quantitative measurement of donor-derived cell-free DNA (cfDNA) would have independent value for the assessment of graft integrity, including damage from acute rejection.
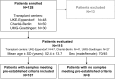
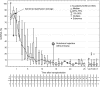
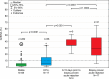
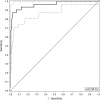
Methods and findings: Traditional LFTs were performed and plasma GcfDNA was monitored in 115 adults post-LTx at three German transplant centers as part of a prospective, observational, multicenter cohort trial. GcfDNA percentage (graft cfDNA/total cfDNA) was measured using droplet digital PCR (ddPCR), based on a limited number of predefined single nucleotide polymorphisms, enabling same-day turn-around. The same method was used to quantify blood microchimerism. GcfDNA was increased >50% on day 1 post-LTx, presumably from ischemia/reperfusion damage, but rapidly declined in patients without graft injury within 7 to 10 d to a median <10%, where it remained for the 1-y observation period. Of 115 patients, 107 provided samples that met preestablished criteria. In 31 samples taken from 17 patients during biopsy-proven acute rejection episodes, the percentage of GcfDNA was elevated substantially (median 29.6%, 95% CI 23.6%-41.0%) compared with that in 282 samples from 88 patients during stable periods (median 3.3%, 95% CI 2.9%-3.7%; p < 0.001). Only slightly higher values (median 5.9%, 95% CI 4.4%-10.3%) were found in 68 samples from 17 hepatitis C virus (HCV)-positive, rejection-free patients. LFTs had low overall correlations (r = 0.28-0.62) with GcfDNA and showed greater overlap between patient subgroups, especially between acute rejection and HCV+ patients. Multivariable logistic regression modeling demonstrated that GcfDNA provided additional LFT-independent information on graft integrity. Diagnostic sensitivity and specificity were 90.3% (95% CI 74.2%-98.0%) and 92.9% (95% CI 89.3%-95.6%), respectively, for GcfDNA at a threshold value of 10%. The area under the receiver operator characteristic curve was higher for GcfDNA (97.1%, 95% CI 93.4%-100%) than for same-day conventional LFTs (AST: 95.7%; ALT: 95.2%; γ-GT: 94.5%; bilirubin: 82.6%). An evaluation of microchimerism revealed that the maximum donor DNA in circulating white blood cells was only 0.068%. GcfDNA percentage can be influenced by major changes in host cfDNA (e.g., due to leukopenia or leukocytosis). One limitation of our study is that exact time-matched GcfDNA and LFT samples were not available for all patient visits.
Conclusions: In this study, determination of GcfDNA in plasma by ddPCR allowed for earlier and more sensitive discrimination of acute rejection in LTx patients as compared with conventional LFTs. Potential blood microchimerism was quantitatively low and had no significant influence on GcfDNA value. Further research, which should ideally include protocol biopsies, will be needed to establish the practical value of GcfDNA measurements in the management of LTx patients.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: ES and JBe are employees of, and own stock and intellectual property rights at Chronix Biomedical, which provided parts of the reagents. MO acts as consultant and advisor to Chronix Biomedical. MK has received research grants from, and honoraria for speaking at Novartis and Teva. PDW was previously been a paid consultant to Chronix Biomedical. MH has served two paid internships in 2013 and 2014 at the Novartis Pharma AG. MH's position at the university was partly funded by the research project the analyses are based on (Federal Ministry of Education and Research (BMBF): "Personalized Immunosuppression after Liver Transplantation: an observational study on the utility and therapeutic ranges of innovative biomarkers"). MH has also received grants from the Federal Ministry of Education and Research for other research projects.
Figures
References
-
- Burton JR Jr, Rosen HR. Acute rejection in HCV-infected liver transplant recipients: the great conundrum. Liver Transpl. 2006;12(11 Suppl 2):S38–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

