Biomonitoring of marine vertebrates in Monterey Bay using eDNA metabarcoding
- PMID: 28441466
- PMCID: PMC5404852
- DOI: 10.1371/journal.pone.0176343
Biomonitoring of marine vertebrates in Monterey Bay using eDNA metabarcoding
Abstract
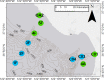
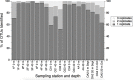
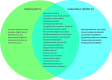
Molecular analysis of environmental DNA (eDNA) can be used to assess vertebrate biodiversity in aquatic systems, but limited work has applied eDNA technologies to marine waters. Further, there is limited understanding of the spatial distribution of vertebrate eDNA in marine waters. Here, we use an eDNA metabarcoding approach to target and amplify a hypervariable region of the mitochondrial 12S rRNA gene to characterize vertebrate communities at 10 oceanographic stations spanning 45 km within the Monterey Bay National Marine Sanctuary (MBNMS). In this study, we collected three biological replicates of small volume water samples (1 L) at 2 depths at each of the 10 stations. We amplified fish mitochondrial DNA using a universal primer set. We obtained 5,644,299 high quality Illumina sequence reads from the environmental samples. The sequence reads were annotated to the lowest taxonomic assignment using a bioinformatics pipeline. The eDNA survey identified, to the lowest taxonomic rank, 7 families, 3 subfamilies, 10 genera, and 72 species of vertebrates at the study sites. These 92 distinct taxa come from 33 unique marine vertebrate families. We observed significantly different vertebrate community composition between sampling depths (0 m and 20/40 m deep) across all stations and significantly different communities at stations located on the continental shelf (<200 m bottom depth) versus in the deeper waters of the canyons of Monterey Bay (>200 m bottom depth). All but 1 family identified using eDNA metabarcoding is known to occur in MBNMS. The study informs the implementation of eDNA metabarcoding for vertebrate biomonitoring.
Conflict of interest statement
Figures

References
-
- Roberts CM, Hawkins JP. Extinction risk in the sea. TREE. 1999;14: 241–246. - PubMed
-
- Kelly RP, Port JA, Yamahara KM, Crowder LB. Using Environmental DNA to Census Marine Fishes in a Large Mesocosm. Hofmann GE, editor. PLoS ONE. 2014;9: e86175–11. doi: 10.1371/journal.pone.0086175 - DOI - PMC - PubMed
-
- McCauley DJ, Pinsky ML, Palumbi SR, Estes JA, Joyce FH, Warner RR. Marine defaunation: Animal loss in the global ocean. Science. 2015;347: 1255641–1255641. doi: 10.1126/science.1255641 - DOI - PubMed
-
- Coll M, Libralato S, Tudela S, Palomera I, Pranovi F. Ecosystem Overfishing in the Ocean. Hector A, editor. PLoS ONE. Public Library of Science; 2008;3: e3881–10. doi: 10.1371/journal.pone.0003881 - DOI - PMC - PubMed
-
- Haigh R, Ianson D, Holt CA, Neate HE, Edwards AM. Effects of Ocean Acidification on Temperate Coastal Marine Ecosystems and Fisheries in the Northeast Pacific. Thiyagarajan Rajan V, editor. PLoS ONE. 2015;10: e0117533–46. doi: 10.1371/journal.pone.0117533 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

