Caenorhabditis elegans PRMT-7 and PRMT-9 Are Evolutionarily Conserved Protein Arginine Methyltransferases with Distinct Substrate Specificities
- PMID: 28441492
- PMCID: PMC5503151
- DOI: 10.1021/acs.biochem.7b00283
Caenorhabditis elegans PRMT-7 and PRMT-9 Are Evolutionarily Conserved Protein Arginine Methyltransferases with Distinct Substrate Specificities
Abstract
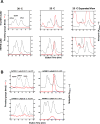
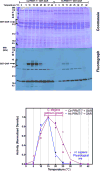
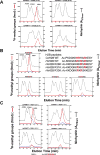
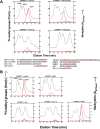
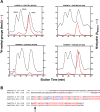
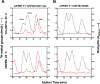
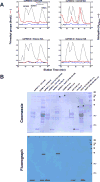
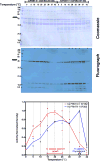
Caenorhabditis elegans protein arginine methyltransferases PRMT-7 and PRMT-9 are two evolutionarily conserved enzymes, with distinct orthologs in plants, invertebrates, and vertebrates. Biochemical characterization of these two enzymes reveals that they share much in common with their mammalian orthologs. C. elegans PRMT-7 produces only monomethylarginine (MMA) and preferentially methylates R-X-R motifs in a broad collection of substrates, including human histone peptides and RG-rich peptides. In addition, the activity of the PRMT-7 enzyme is dependent on temperature, the presence of metal ions, and the reducing agent dithiothreitol. C. elegans PRMT-7 has a substrate specificity and a substrate preference different from those of mammalian PRMT7, and the available X-ray crystal structures of the PRMT7 orthologs show differences in active site architecture. C. elegans PRMT-9, on the other hand, produces symmetric dimethylarginine and MMA on SFTB-2, the conserved C. elegans ortholog of human RNA splicing factor SF3B2, indicating a possible role in the regulation of nematode splicing. In contrast to PRMT-7, C. elegans PRMT-9 appears to be biochemically indistinguishable from its human ortholog.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Wang Y, Wang J, Chen C, Chen Y, Li C. A novel BLAST-Based Relative Distance (BBRD) method can effectively group members of protein arginine methyltransferases and suggest their evolutionary relationship. Mol Phylogenet Evol. 2015;84:101–111. - PubMed
-
- Wang YC, Li C. Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 2012;279:932–945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

