Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes
- PMID: 28441772
- PMCID: PMC6154101
- DOI: 10.3390/molecules22040678
Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes
Abstract
Fourteen well-defined ribozyme classes have been identified to date, among which nine are site-specific self-cleaving ribozymes. Very recently, small self-cleaving ribozymes have attracted renewed interest in their structure, biochemistry, and biological function since the discovery, during the last three years, of four novel ribozymes, termed twister, twister sister, pistol, and hatchet. In this review, we mainly address the structure, biochemistry, and catalytic mechanism of the novel ribozymes. They are characterized by distinct active site architectures and divergent, but similar, biochemical properties. The cleavage activities of the ribozymes are highly dependent upon divalent cations, pH, and base-specific mutations, which can cause changes in the nucleotide arrangement and/or electrostatic potential around the cleavage site. It is most likely that a guanine and adenine in close proximity of the cleavage site are involved in general acid-base catalysis. In addition, metal ions appear to play a structural rather than catalytic role although some of their crystal structures have shown a direct metal ion coordination to a non-bridging phosphate oxygen at the cleavage site. Collectively, the structural and biochemical data of the four newest ribozymes could contribute to advance our mechanistic understanding of how self-cleaving ribozymes accomplish their efficient site-specific RNA cleavages.
Keywords: catalytic mechanism; hatchet; novel ribozymes; pistol; structure; twister; twister-sister.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

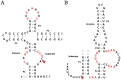
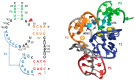
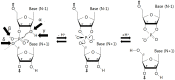
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

