Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome
- PMID: 28441964
- PMCID: PMC5405467
- DOI: 10.1186/s40168-017-0261-y
Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome
Abstract
Background: Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is characterized by unexplained persistent fatigue, commonly accompanied by cognitive dysfunction, sleeping disturbances, orthostatic intolerance, fever, lymphadenopathy, and irritable bowel syndrome (IBS). The extent to which the gastrointestinal microbiome and peripheral inflammation are associated with ME/CFS remains unclear. We pursued rigorous clinical characterization, fecal bacterial metagenomics, and plasma immune molecule analyses in 50 ME/CFS patients and 50 healthy controls frequency-matched for age, sex, race/ethnicity, geographic site, and season of sampling.
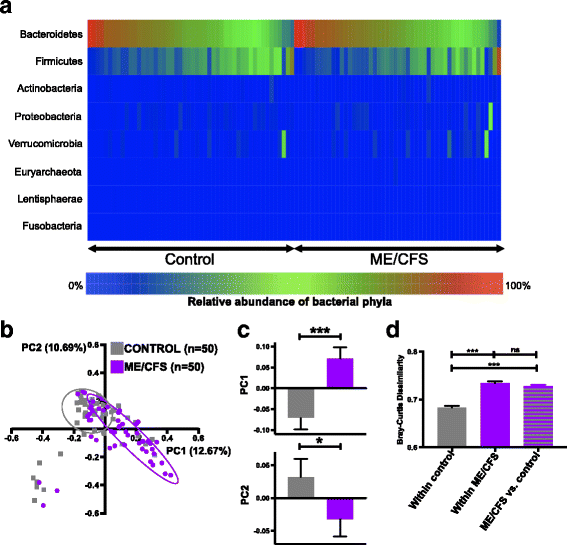
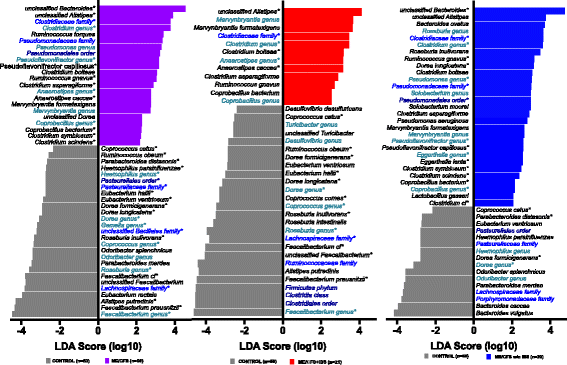
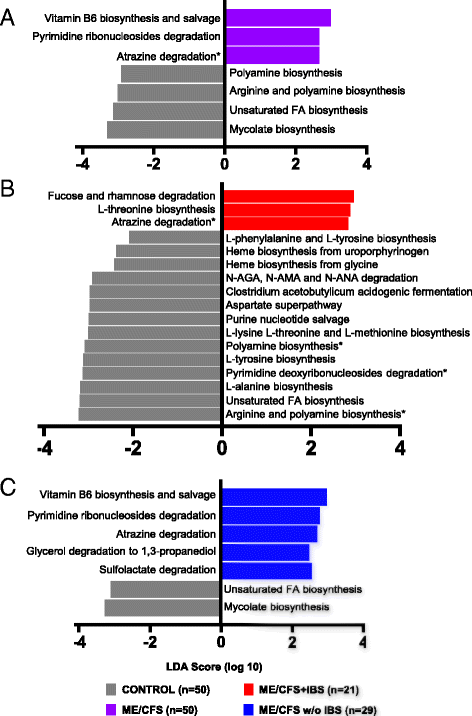
Results: Topological analysis revealed associations between IBS co-morbidity, body mass index, fecal bacterial composition, and bacterial metabolic pathways but not plasma immune molecules. IBS co-morbidity was the strongest driving factor in the separation of topological networks based on bacterial profiles and metabolic pathways. Predictive selection models based on bacterial profiles supported findings from topological analyses indicating that ME/CFS subgroups, defined by IBS status, could be distinguished from control subjects with high predictive accuracy. Bacterial taxa predictive of ME/CFS patients with IBS were distinct from taxa associated with ME/CFS patients without IBS. Increased abundance of unclassified Alistipes and decreased Faecalibacterium emerged as the top biomarkers of ME/CFS with IBS; while increased unclassified Bacteroides abundance and decreased Bacteroides vulgatus were the top biomarkers of ME/CFS without IBS. Despite findings of differences in bacterial taxa and metabolic pathways defining ME/CFS subgroups, decreased metabolic pathways associated with unsaturated fatty acid biosynthesis and increased atrazine degradation pathways were independent of IBS co-morbidity. Increased vitamin B6 biosynthesis/salvage and pyrimidine ribonucleoside degradation were the top metabolic pathways in ME/CFS without IBS as well as in the total ME/CFS cohort. In ME/CFS subgroups, symptom severity measures including pain, fatigue, and reduced motivation were correlated with the abundance of distinct bacterial taxa and metabolic pathways.
Conclusions: Independent of IBS, ME/CFS is associated with dysbiosis and distinct bacterial metabolic disturbances that may influence disease severity. However, our findings indicate that dysbiotic features that are uniquely ME/CFS-associated may be masked by disturbances arising from the high prevalence of IBS co-morbidity in ME/CFS. These insights may enable more accurate diagnosis and lead to insights that inform the development of specific therapeutic strategies in ME/CFS subgroups.
Keywords: Chronic fatigue syndrome; Irritable bowel syndrome; Metabolic pathway; Metagenomic; Microbiota-gut-brain axis; Myalgic encephalomyelitis; Topological data analysis.
Figures


References
-
- Institute of Medicine I . Beyond myalgic encephalomyelitis/chronic fatigue syndrome: redefining an illness. Washington: The National Academies Press; 2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

